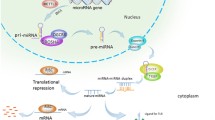
Abstract
MicroRNAs (miRNAs) are members of the non-protein coding RNA family. miRNAs, which can regulate genes on transcriptomic level through either degrading the target messenger RNA (mRNA) or suppressing the protein synthesis, also take part in a number of biological functions that involve development, differentiation, proliferation and apoptosis. The mutations and polymorphisms in the expression levels of miRNA genes or alterations in their epigenetic mechanisms may play their roles in the formation of malignancies. Increasing evidence shows that aberrant miRNA expression profiles are present in a variety of cancers. Therefore, it has been suggested that these profiles could be useful for diagnosis and classification of different tumor types and that these small RNAs might provide significant opportunities for the development of future miRNA-based therapies. In this review, we aimed to look into the realm of miRNAs, which is a recent area of research, appraise their biological activities on molecular level and their probable benefits on clinical practice.

Similar content being viewed by others
References
Lee RC, Feinbaum RC, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with anti-sense complementary to lin-4. Cell 75(5):843–854
Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP (2003) Vertebrata microRNA genes. Science 299(5612):1540
Lynam-Lennon N, Maher SG, Reynolds JV (2009) The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc 84(1):55–71
Yendamuri S, Calin GA (2009) The role of microRNA in human leukemia: a review. Leukemia 23(7):1257–1263
miRBase. Available from: http://www.mirbase.org/. Accessed 4 June 2010
Wang V, Wu W (2009) MicroRNA-based therapeutics for cancer. BioDrugs 23(1):15–23
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM (2002) Frequent deletions and down-regulation of micro-RNA genes miR-15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99(24):15524–15529
Rana TM (2007) Illuminating the science: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 8(1):23–36
Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10(10):704–714
Fabbri M (2008) MicroRNAs and cancer epigenetics. Curr Opin Investig Drugs 9(6):583–590
Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10(2):126–139
Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6(5):376–385
Borchert GM, Lanier W, Davidson BL (2006) RNA polymerase III transcribes human microRNA. Nat Struct Mol Biol 13(12):1097–1101
Pawlicki JM, Steitz JA (2008) Primary microRNA transcript retention at sites of transcripsion leads to enhanced microRNA production. J Cell Biol 182(1):61–76
Kim YK, Kim VN (2007) Processing of intronic microRNAs. EMBO J 26(3):775–783
Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130(1):89–100
Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448(7149):83–86
Cowland JB, Hother C, Grønbaek K (2007) MicroRNAs and cancer. APMIS 115(10):1090–1106
Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W (2008) Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis 29(7):1306–1311
Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9(3):219–230
Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9(1):22–32
Wilshem JE, Smibert CA (2005) Mechanisms of translational regulation in Drosophila. Biol Cell 97(4):235–252
Standart N, Minshall N (2008) Translational control in early development: CPEB, P-bodies and germinal granules. Biochem Soc Trans 36(Pt 4):671–676
Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447(7146):823–828
Wang M, Tan LP, Dijkstra MK, Van Lom K, Robertus JL, Harms G, Blokzijl T, Kooistra K, van T’veer MB, Rosati S, Visser L, Jongen-Lavrencic M, Kluin PM, van den Berg A (2008) miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J Pathol 215(1):13–20
Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P-bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8(1):9–22
Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40(1):43–50
He L, He X, Lowe SW, Hannon GJ (2007) microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer 7(11):819–822
He L, He X, Lim LP, De Stanchine E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ (2007) A microRNA component of the p53 tumour suppressor network. Nature 447(7148):1130–1134
Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I (2005) A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBP alpha regulates human granulopoiesis. Cell 123(5):819–831
Barbarotto E, Schmittgen TD, Calin GA (2008) MicroRNAs and cancer: profile, profile, profile. Int J Cancer 122(5):969–977
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435(7043):828–833
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM (2004) Human microRNA genes are frequently located at frajile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101(9):2999–3004
Baffa R, Fassan M, Volinia S, O’Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, Rosenberg A (2009) MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol 219(2):214–221
Xu W, Li JY (2007) MicroRNA gene expression in malignant Iymphoproliferative disorders. Chin Med J 120(11):996–999
Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M (2008) Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR–ABL1 oncogene expression. Cancer Cell 13(6):496–506
Faber J, Gregory RI, Armstrong SA (2008) Linking miRNA regulation to BCR–ABL expression: the next dimension. Cancer Cell 13(6):467–469
Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 104(40):15805–15810
Duursma AM, Kedde M, Schrier M, Le Sage C, Agami R (2008) miR-148 targets human DNMT3b protein coding region. RNA 14(5):872–877
Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38(2):228–233
Benetti R, Gonzalo S, Jaco I, Muňos P, Gonzales S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA (2008) A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl-2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15(3):268–279
Misha PJ, Bertino JR (2009) MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 10(3):399–416
Gottwein E, Cai X, Cullen BR (2006) A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol 80(11):5321–5326
Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T (2007) Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39(5):673–677
Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, Toro JR, Zenger VE, Metcalf RA, Marti GE (2007) Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood 109(12):5079–5086
Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM (2005) A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353(17):1793–1801
Duan R, Pak C, Jin P (2007) Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 16(9):1124–1131
Misha PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GSA, Banerjee D, Bertino J (2007) A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA 104(33):13513–13518
Tchatchou S, Jung A, Hemminki K, Sutter C, Wappenschmidt B, Bugert P, Weber BH, Niederacher D, Arnold N, Varon-Mateeva R, Ditsch N, Meindl A, Schmutzler RK, Bartram CR, Burwinkel B (2009) A variant affecting a putative miRNA target site in estrogen receptor (ESR) 1 is associated with breast cancer risk in premenopausal women. Carcinogenesis 30(1):59–64
Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, Marsit CJ, Kelsey KT (2009) A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis 30(6):1003–1007
Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB (2008) A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 68(20):8535–8540
Sassen S, Miska EA, Caldas C (2008) MicroRNA: implications for cancer. Virchows Arch 452(1):1–10
He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A (2005) The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 102(52):19075–19080
Lv K, Guo Y, Zhang Y, Wang K, Jia Y, Sun S (2008) Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/deletion polymorphism with type 2 diabetes. Biochem Biophys Res Commun 374(1):101–105
Passetti F, Ferreira CG, Costa FF (2009) The impact of microRNAs and alternative splicing in pharmacogenomics. Pharmacogenomics J 9(1):1–13
Saunders MA, Liang H, Li WH (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA 104(9):3300–3305
Iwai N, Naraba H (2005) Polymorphisms in human pre-miRNAs. Biochem Biophys Res Commun 331(4):1439–1444
Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A (2009) Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA 106(5):1502–1505
Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H (2008) A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis 29(10):1963–1966
Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 105(20):7269–7274
Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M (2008) A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA 105(36):13556–13561
Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27(15):2128–2136
Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18(3):350–359
Huang Q, Gumireddy K, Schrier M, Le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Puré E, Agami R (2008) The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 10(2):202–210
Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E (2009) MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 125(6):1407–1413
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449(7163):682–688
Negrini M, Calin GA (2008) Breast cancer metastasis: a microRNA story. Breast Cancer Res 10(2):203
Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451(7175):147–152
Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY (2009) p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA 106(9):3207–3212
Kuehbacher A, Urbich C, Dimmeler S (2008) Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci 29(1):12–15
Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y (2006) MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1:e116
Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G (2006) MicroRNAs modulate the angiogenic properties of HUVECs. Blood 108(9):3068–3071
Suárez Y, Fernández-Hernando C, Pober JS, Sessa WC (2007) Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 100(8):1164–1173
Guo CJ, Pan Q, Li DG, Sun H, Liu BW (2009) miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: an essential role for apoptosis. J Hepatol 50(4):766–778
Akao Y, Nakagawa Y, Naoe T (2006) MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep 16(4):845–850
Pekarsky Y, Calin GA, Aqeilan R (2005) Chronic lymphocytic leukemia: molecular genetics and animal models. Curr Top Microbiol Immunol 294:51–70
Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA (2006) Specific activation of microRNA-127 with downregulation of proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9(6):435–443
Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y (2008) MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27(31):4373–4379
Pedersen IM, Otero D, Kao E, Miletic AV, Hother C, Ralfkiaer E, Rickert RC, Gronbaek K, David M (2009) Onco-miR-155 targets SHIP1 to promote TNFa-dependent growth of B cell lymphomas. EMBO Mol Med 1(5):288–295
Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG (2005) Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334(4):1351–1358
Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C (2005) MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA 102(50):18081–18086
Lehmann U, Hasemeier B, Christgen M, Müller M, Römermann D, Länger F, Kreipe H (2008) Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol 214(1):17–24
Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, Teh BT (2009) MicroRNA profiling of human kidney cancer subtypes. Int J Oncol 35(1):109–114
Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R (2007) Micro-RNA profiling in kidney and bladder cancers. Urol Oncol 25(5):387–392
McLaughlin J, Cheng D, Singer O, Lukacs RU, Radu CG, Verma IM, Witte ON (2007) Sustained suppression of Bcr-Abl-driven lymphoid leukemia by microRNA mimics. Proc Natl Acad Sci USA 104(51):20501–20506
Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY (2007) miR-21-mediated tumor growth. Oncogene 26(19):2799–2803
Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438(7068):685–689
Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4(9):721–726
Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M (2004) Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res 64(9):3087–3095
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103(7):2257–2261
Pierson J, Hostager B, Fan R, Vibhakar R (2008) Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 90(1):1–7
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Varol, N., Konac, E., Gurocak, O.S. et al. The realm of microRNAs in cancers. Mol Biol Rep 38, 1079–1089 (2011). https://doi.org/10.1007/s11033-010-0205-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0205-0