Abstract
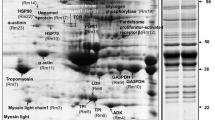
Protein profile of the skeletal muscle of Takifugu rubripes, a kind of pufferfish, was carried out with two-dimensional polyacrylamide gel electrophoresis (2-DE). Among the 112 protein spots detected in a silver-stained 2-D polyacrylamide gel, 33 were analyzed by Matrix Assisted Laser Desorption Ionisation tandem time-of-flight mass spectrometry (MALDI TOF/TOF MS), and 21 were identified by MASCOT. There were six structural proteins, such as alpha-actin, tropomyosin, and myosin heavy chain, and six with known functions such as T-cell receptor alpha chain, 4SNc-Tudor domain protein, SMC3 protein, and Translin associated factor X, as well as nine hypothetical novel proteins, including titin, andretinol dehydrogenase, and apolipoprotein A-I binding protein. These proteins were further categorized into six functional groups. This paper established a suitable technical protocol to eliminate the high abundance proteins while preserving middle abundance proteins for proteomics studies using Takifugu skeletal muscle. It is also favorable for further investigation on screening marker proteins for monitoring and controlling the quality of fish meat.






Similar content being viewed by others
References
Brenner S, Elgar G, Sandford R et al (1993) Characterisation of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366:265–268. doi:10.1038/366265a0
Aparicio S, Chapman J, Stupka E et al (2002) Whole genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310. doi:10.1126/science.1072104
Carbonaro M (2004) Proteomics: present and future in food quality evaluation. Trends Food Sci Technol 15:209–216. doi:10.1016/j.tifs.2003.09.020
Lametsch R, Bendixen E (2001) Proteome analysis applied to meat science: characterizing post mortem changes in porcine muscle. J Agric Food Chem 49:4531–4537. doi:10.1021/jf010103g
Doherty MK, McLean L, Hayter JR (2004) The proteome of chicken skeletal muscle: changes in soluble protein expression during growth in a layer strain. Proteomics 4:2082–2093. doi:10.1002/pmic.200300716
Bendixen E (2005) The use of proteomics in meat science. Meat Sci 71:138–149. doi:10.1016/j.meatsci.2005.03.013
Westermeier R, Naven T (2002) Proteomics in practice: a laboratory manual of proteome analysis practical proteomics. Wiley-VCH, Weinheim, Germany
Kuan W, Janela M, Ann T (1979) Titin: major myofibrillar components of striated muscle. Proc Natl Acad Sci USA 76:3698–3702. doi:10.1073/pnas.76.8.3698
Fürst DO, Osborn M, Weber K (1989) Myogenesis in the mouse embryo: differential onset of expression of myogenic proteins and the involvement of titin in myofibril assembly. J Cell Biol 109:517–527. doi:10.1083/jcb.109.2.517
Whiting A, Wardale J, Trinick J (1989) Does titin regulate the length of muscle thick filaments? J Mol Biol 205:163–169. doi:10.1016/0022-2836(89)90381-1
Henk LG, Siegfried L (2004) The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ Res 94:284–295. doi:10.1161/01.RES.0000117769.88862.F8
Yoshida Y, Kojima N, Kurosawa N (1995) Molecular cloning of Sia alpha-2, 3-Gal-beta 1, 4-Glc-Nac-alpha 2, 8-sialyltransferase from mouse brain. J Biol Chem 270:14628–14633. doi:10.1074/jbc.270.24.14628
Patrick JB, Christine T, Michele D (1997) Both apolipoprotein E and A-I genes are present in a nonmammalian vertebrate and are highly expressed during embryonic development. Proc Natl Acad Sci USA 94:8622–8627. doi:10.1073/pnas.94.16.8622
Usener D, Schadendorf D, Koch J, Dübel S, Eichmüller S (2003) cTAGE: a cutaneous T cell lymphoma associated antigen family with tumor-specific splicing. Invest Dermatol 121:198–206. doi:10.1046/j.1523-1747.2003.12318.x
Giancarlo G (2006) SMC3 knockdown triggers genomic instability and p53-dependent apoptosis in human and zebrafish cells. Mol Cancer 5:52
Watrin E, Peters JM (2006) Cohesin and DNA damage repair. Exp Cell Res 312(14):2687–2693. doi:10.1016/j.yexcr.2006.06.024
Deardorff MA, Kaur M, Yaeger D et al (2007) Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 80(3):485–494. doi:10.1086/511888
Tuba E, Bilada B, Dilhan O et al (2002) DNA damage-dependent interaction of the nuclear matrix protein C1D with translin-associated factor X (TRAX). J Cell Sci 115:207–216
Chennathukuzhi VM, Kurihara Y, Bray JD et al (2001) Trax (Translin-associated Factor X), a primarily cytoplasmic protein, inhibits the binding of TB-RBP (Translin) to RNA. J Biol Chem 276:13256–13263. doi:10.1074/jbc.M009707200
Tong X, Drapkin R, Yalamanchili R et al (1995) The Epstein-Barr virus nuclear protein acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol 15:4735–4744
Ponting CP (1997) Tudor domains in proteins that interact with RNA. Trends Biochem Sci 2:51–52. doi:10.1016/S0968-0004(96)30049-2
Porta A, Colonna-Romano S, Callebaut I et al (1999) A homologue of the human 100 kDa protein (p100) is differentially expressed by Histoplasma capsulatum during infection of murine macrophages. Biochem Biophys Res Commun 254:605–613. doi:10.1006/bbrc.1998.9894
Shunnosuke A, Masako S, Kosaku Y, Takehiko H, Katsumasa O, Koichi S, Eric D (2002) A Tudor protein with multiple SNc domains from pea seedlings: cellular localization, partial characterization, sequence analysis, and phylogenetic relationships. J Exp Bot 384:971–983
Li C-L, Yang W-Z, Chen Y-P et al (2008) Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res 36(11):3579–3589. doi:10.1093/nar/gkn236
Alexander D, Andreas M (2002) Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol 159:255–266. doi:10.1083/jcb.200204023
Acknowledgments
This work was supported by Natural Science Fund for Colleges and Universities in Jiangsu Province, China (No. 08KJD180008). We thank Professor Yong Wang (Jiangsu University, China) for his revision of our manuscript.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lu, J., Zheng, J., Liu, H. et al. Protein profiling analysis of skeletal muscle of a pufferfish, Takifugu rubripes . Mol Biol Rep 37, 2141–2147 (2010). https://doi.org/10.1007/s11033-009-9684-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9684-2