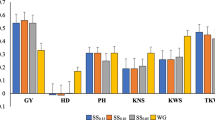
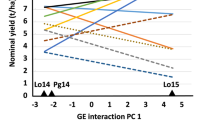
Abstract
Germplasm collections hold several thousands of white lupin (Lupinus albus L.) accessions. Genome-enabled models with good predictive ability for specific environments could provide a cost-efficient means to identify promising genetic resources for breeding programmes. This study provided an unprecedented assessment of genome-enabled predictions for white lupin grain yield, focusing on (i) a world collection of 109 landraces and 8 varieties phenotyped in three European sites with contrasting climate (Mediterranean, subcontinental or oceanic) and sowing time (data set 1); (ii) 78 geographically diversified landrace genotypes and three variety genotypes phenotyped in moisture-favourable and severely drought-prone managed environments (data set 2). The interest of predictions for individual genotypes was justified by large within-landrace variation for yield responses. Ridge regression BLUP (rrBLUP) and Bayesian Lasso (BL) models exploited allele frequencies (estimated from 3 to 4 genotypes per landrace) of 10,782 polymorphic SNPs for data set 1, and allele values of 9937 polymorphic SNPs for data set 2, following ApeKI-based genotyping-by-sequencing characterization. Compared with BL, rrBLUP displayed similar predictive ability for data set 1 and better predictive ability for data set 2. Best-predictive models displayed intra-environment predictive ability for the five test environments in the range 0.47–0.76. Cross-environment predictions between pairs of environments with positive genetic correlation, i.e., autumn-sown subcontinental vs Mediterranean sites, and moisture-favourable vs drought-prone environments, exhibited a predictive ability range of 0.40–0.51 and a predictive accuracy range of 0.48–0.61. Our results support the exploitation of genomic predictions and provide economic justification for the genotyping of germplasm collections of white lupin.
Similar content being viewed by others
References
Alessandri A, De Felice M, Zeng N, Mariotti A, Pan Y, Cherchi A et al (2014) Robust assessment of the expansion and retreat of Mediterranean climate in the 21st century. Sci Rep 4:7211
Annicchiarico P (2008) Adaptation of cool-season grain legume species across climatically-contrasting environments of southern Europe. Agron J 100:1647–1654
Annicchiarico P (2009) Coping with and exploiting genotype × environment interactions. In: Ceccarelli S, Guimarães EP, Weltzien E (eds) Plant breeding and farmer participation. FAO, Rome, pp 519–564
Annicchiarico P, Carroni AM (2009) Diversity of white and narrow-leafed lupin genotype adaptive response across south-European environments and implications for selection. Euphytica 166:71–81
Annicchiarico P, Harzic N, Carroni AM (2010) Adaptation, diversity and exploitation of global white lupin (Lupinus albus L.) landrace genetic resources. Field Crops Res 119:114–124
Annicchiarico P, Harzic N, Huyghe C, Carroni AM (2011) Ecological classification of white lupin landrace genetic resources. Euphytica 180:17–25
Annicchiarico P, Nazzicari N, Li X, Wei Y, Pecetti L, Brummer EC (2015) Accuracy of genomic selection for alfalfa biomass yield in different reference populations. BMC Genomics 16:1020
Annicchiarico P, Nazzicari N, Pecetti L, Romani M, Ferrari B, Wei Y, et al. (2017a) GBS-based genomic selection for pea grain yield under severe terminal drought. Plant Genome 10:2
Annicchiarico P, Nazzicari N, Wei Y, Pecetti L, Brummer EC (2017b) Genotyping-by-sequencing and its exploitation for forage and cool-season grain legume breeding. Front Plant Sci 8:679
Annicchiarico P, Romani M, Pecetti L (2018) White lupin variation for adaptation to severe drought stress. Plant Breed 137:782–789
Annicchiarico P, Nazzicari N, Pecetti L, Romani M, Russi L (2019) Pea genomic selection for Italian environments. BMC Genomics 20:603
Arnoldi A, Boschin G, Zanoni C, Lammi C (2015) The health benefits of sweet lupin seed flours and isolated proteins. J Funct Foods 18:550–563
Atkins CA, Smith PMC, Gupta S, Jones MGK, Caligari PDS (1998) Genetics, cytology and biotechnology. In: Gladstones JS, Atkins C, Hamblin J (eds) Lupins as crop plants: biology, production and utilization. CAB International, Wallingford, UK, pp 67–92
Atnaf M, Yao N, Martina K, Dagne K, Wegary D, Tesfaye K (2017) Molecular genetic diversity and population structure of Ethiopian white lupin landraces: implications for breeding and conservation. PLoS One 12:e0188696
Bassi FM, Bentley AR, Charmet G, Ortiz R, Crossa J (2016) Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci 242:23–36
Boschin G, D’Agostina A, Annicchiarico P, Arnoldi A (2007) The fatty acid composition of the oil from Lupinus albus cv. Luxe as affected by environmental and agricultural factors. Eur Food Res Technol 225:769–776
Breiman L (2001) Random forests. Mach Learn 45:5–32
Brown AHD (2000) The genetic structure of crop landraces and the challenge to conserve them in situ on farms. In: Brush SB (ed) Genes in the field. On-farm conservation of crop diversity. IPGRI/IDRC/Lewis Publishers, Boca Raton, FL, pp 19–48
Buirchell BJ, Cowling WA (1998) Genetic resources in lupins. In: Gladstones JS, Atkins C, Hamblin J (eds) Lupins as crop plants: biology, production and utilization. CAB International, Wallingford, UK, pp 41–66
Burstin J, Salloignon P, Chabert-Martinello M, Magnin-Robert J-B, Siol M, Jacquin F et al. (2015) Genetic diversity and trait genomic prediction in a pea diversity panel. BMC Genomics 16:105
Casella G, George EI (1992) Explaining the Gibbs sampler. Am Stat 46:167–174
Chong Z, Ruan J, Wu CI (2012) Rainbow: an integrated tool for efficient clustering and assembling RAD-seq reads. Bioinformatics 28:2732–2737
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
DeLacy IH, Basford KE, Cooper M, Bull IK, McLaren CG (1996) Analysis of multi-environment trials – an historical perspective. In: Cooper M, Hammer GL (eds) Plant adaptation and crop improvement. CAB International, Wallingford, UK, pp 39–124
Duhnen A, Gras A, Teyssèdre S, Romestant M, Claustres B, Daydé J, et al. (2017) Genomic selection for yield and seed protein content in soybean: a study of breeding program data and assessment of prediction accuracy. Crop Sci 57:1325–1337
Elbasyoni IS, Lorenz AJ, Guttieri M, Frels K, Baenziger PS, Poland J, et al. (2018) A comparison between genotyping-by-sequencing and array-based scoring of SNPs for genomic prediction accuracy in winter wheat. Plant Sci 270:123–130
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379
Gladstones JS (1998) Distribution, origin, taxonomy, history and importance. In: Gladstones JS, Atkins C, Hamblin J (eds) Lupins as crop plants: biology, production and utilization. CAB International, Wallingford, UK, pp 1–39
Gresta F, Wink M, Prins U, Abberton M, Capraro J, Scarafoni A, et al. (2017) Lupins in European cropping systems. In: Murphy-Bokern D, Stoddard FL, Watson CA (eds) Legumes in cropping systems. CAB International, Wallingford, UK, pp 88–108
Guo Z, Tucker DM, Basten CJ, Gandhi H, Ersoz E, Guo B et al. (2014) The impact of population structure on genomic prediction in stratified populations. Theor Appl Genet 127:749–762
Heffner EL, Sorrells ME, Jannink J-L (2009) Genomic selection for crop improvement. Crop Sci 49(1):12
Huyghe C (1997) White lupin (Lupinus albus L.). Field Crops Res 53:147–160
Huyghe C, Papineau J (1990) Winter development of autumn-sown white lupin: agronomic and breeding consequences. Agronomie 10:709–716
Huyghe C, Julier B, Harzic N, Papineau J (1994) Yield and yield components of indeterminate autumn-sown white lupin (Lupinus albus) cv. Lunoble. Eur J Agron 3:145–152
Jarquín D, Kocak K, Posadas L, Hyma K, Jedlicka J, Graef G, et al. (2014) Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genomics 15:740
Jarquín D, Specht J, Lorenz A (2016) Prospects of genomic prediction in the USDA soybean germplasm collection: historical data creates robust models for enhancing selection of accessions. G3 (Bethesda) 6:2329–2341
Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, et al. (2008) Efficient control of population structure in model organism association mapping. Genetics 178:1709–1723
Książkiewicz M, Nazzicari N, Yang H, Nelson N, Renshaw D, Rychel S et al (2017) A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci Rep 7:15335
Li X, Wei Y, Acharya A, Hansen JL, Crawford JL, Viands et al. (2015) Genomic prediction of biomass yield in two selection cycles of a tetraploid alfalfa breeding population. Plant Genome 8:2
Lorenz AJ, Chao S, Asoro FG, Heffner EL, Hayashi T, Iwata H et al (2011) Genomic selection in plant breeding: knowledge and prospects. Adv Agron 110:77–123
Ma Y, Reif JC, Jiang Y, Wen Z, Wang D, Liu Z et al. (2016) Potential of marker selection to increase prediction accuracy of genomic selection in soybean (Glycine max L.). Mol Breed 36:113
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mousavi-Derazmahalleh M, Bayer PE, Nevado B, Hurgobin B, Filatov D, Kilian A et al. (2018) Exploring the genetic and adaptive diversity of a pan-Mediterranean crop wild relative: narrow-leafed lupin. Theor Appl Genet 131:887–901
Nazzicari N, Biscarini F (2017) GROAN: genomic regression workbench (version 1.0.0). https://cran.r-project.org/package=GROAN. Accessed 30 April 2019
Nazzicari N, Biscarini F, Cozzi P, Brummer EC, Annicchiarico P (2016) Marker imputation efficiency for genotyping-by-sequencing data in rice (Oryza sativa) and alfalfa (Medicago sativa). Mol Breed 36:69
Nizam Uddin M, Ellison FW, O’Brien L, Latter BDH (1994) The performance of pure lines derived from heterotic bread wheat hybrids. Aust J Agric Res 45:591–600
Papineau J, Huyghe C (2004) Le lupin doux protéagineux. Editions France Agricole, Paris
Park T, Casella G (2008) The Bayesian Lasso. J Am Stat Assoc 103:681–686
Puritz JB, Hollenbeck CM, Gold JR (2014) dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ 2:e431
Rajsic P, Weersink A, Navabi A, Pauls KP (2016) Economics of genomic selection: the role of prediction accuracy and relative genotyping costs. Euphytica 210:259–276
Riedelsheimer C, Technow F, Melchinger AE (2012) Comparison of whole-genome prediction models for traits with contrasting genetic architecture in a diversity panel of maize inbred lines. BMC Genomics 13:452
Robertson A (1959) The sampling variance of the genetic correlation coefficient. Biometrics 15:469–485
Roorkiwal M, Rathore A, Das RR, Singh MK, Jain A, Srinivasan S et al (2016) Genome-enabled prediction models for yield related traits in chickpea. Front Plant Sci 7:1666
SAS Institute (2011) SAS/STAT® 9.3 User's guide. SAS Institute Inc, Cary, NC
Searle SR, Casella G, McCulloch CE (2009) Variance components. John Wiley & Sons, New York
Soriano Viana JM, Piepho H-P, Silva FF (2016) Quantitative genetics theory for genomic selection and efficiency of breeding value prediction in open-pollinated populations. Sci Agric 73:243–251
Stekhoven DJ, Bühlmann P (2012) MissForest–non-parametric missing value imputation for mixed-type data. Bioinformatics 28:112–118
Wang X, Xu Y, Hu Z, Xu C (2018) Genomic selection methods for crop improvement: current status and prospects. Crop J 6:330–340
Wiggans GR, Cole JB, Hubbard SM, Sonstegard TS (2017) Genomic selection in dairy cattle: the USDA experience. Ann Rev Anim Biosci 5:309–327
Acknowledgements
We are grateful to the Elshire Group Ltd. for the excellent GBS genotyping service and to A. Passerini, P. Gaudenzi, S. Proietti, P. Manunza, G. Rochas and N. Rousseau for technical assistance.
Funding
The experiment data for this study were generated by the project ‘Legumes for the agriculture of tomorrow (LEGATO)’ funded by the FP7 of the European Commission (Grant agreement No. 613551) and the projects ‘Increase of protein feed production’ and ‘Plant Genetic Resources - FAO Treaty’ funded by the Italian Ministry of Agricultural, Food and Forestry Policies.
Author information
Authors and Affiliations
Contributions
PA designed and supervised the research work, obtained financial resources, analysed the phenotypic data and drafted the manuscript. NN was responsible for the bioinformatics pipeline and the definition of genome-enabled models. PA, AMC, NH, MR and LP were responsible for phenotyping experiments. BF collected and verified the quality of DNA samples. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Annicchiarico, P., Nazzicari, N., Ferrari, B. et al. Genomic prediction of grain yield in contrasting environments for white lupin genetic resources. Mol Breeding 39, 142 (2019). https://doi.org/10.1007/s11032-019-1048-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1048-6