Abstract
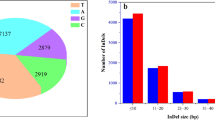
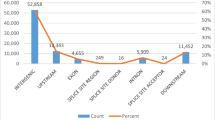
Tobacco (Nicotiana tabacum L., 2n = 48) is an important agronomic crop and model plant. Flue-cured tobacco is the most important type and accounts for approximately 80 % of tobacco production worldwide. The low genetic diversity of flue-cured tobacco impedes the construction of a high-density genetic linkage map using simple sequence repeat (SSR) markers and warrants the exploitation of single nucleotide polymorphic (SNP) markers from genomic regions. In this article, initially using specific locus-amplified fragment sequencing, we discovered 10,891 SNPs that were subsequently used as molecular markers for genetic map construction. Combined with SSR markers, a final high-density genetic map was generated containing 4215 SNPs and 194 SSRs distributed on 24 linkage groups (LGs). The genetic map was 2662.43 cM in length, with an average distance of 0.60 cM between adjacent markers. Furthermore, by mapping the SNP markers to the ancestral genomes of Nicotiana tomentosiformis and Nicotiana sylvestris, a large number of genome rearrangements were identified as occurring after the polyploidization event. Finally, using this novel integrated map and mapping population, two major quantitative trait loci (QTLs) were identified for flue-curing and mapped to the LG6 of tobacco. This is the first report of SNP markers and a SNP-based linkage map being developed in tobacco. The high-density genetic map and QTLs related to tobacco curing will support gene/QTL fine mapping, genome sequence assembly and molecular breeding in tobacco.




Similar content being viewed by others
References
Barchi L, Lanteri S, Portis E, Vale G, Volante A, Pulcini L, Ciriaci T, Acciarri N, Barbierato V, Toppino L, Rotino GL (2012) A RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS One 7(8):e43740. doi:10.1371/journal.pone.0043740
Bindler G, van der Hoeven R, Gunduz I, Plieske J, Ganal M, Rossi L, Gadani F, Donini P (2007) A microsatellite marker based linkage map of tobacco. Theor Appl Genet 114(2):341–349
Bindler G, Plieske J, Bakaher N, Gunduz I, Ivanov N, Van der Hoeven R, Ganal M, Donini P (2011) A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development. Theor Appl Genet 123(2):219–230
Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller L, Martin G (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25:1523–1530
Brenchley R, Spannagl M, Pfeifer M et al (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491(7426):705–710. doi:10.1038/nature11650
Ching A, Caldwell KS, Jung M, Dolan M, Smith OS, Tingey S, Morgante M, Rafalski AJ (2002) SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet. doi:10.1186/1471-2156-3-19
Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol 168(1):241–252. doi:10.1111/j.1469-8137.2005.01480.x
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142(1–2):169–196. doi:10.1007/s10681-005-1681-5
Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42:443–461. doi:10.1146/annurev.genet.42.110807.091524
Gadani F, Hayes A, Opperman CH et al (2003) Large scale genome sequencing and analysis of Nicotiana tabacum: the tobacco genome initiative. In: Proceedings, 5èmes Journées ScientiWques du Tabac de Bergerac—5th bergerac tobacco ScientiWc meeting, Bergerac, pp 117–130. http://www.altadis-bergerac.com/pdf/5_JS_Bergerac.pdf
Kim S, Park M, Yeom SI et al (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46(3):270–278. doi:10.1038/ng.2877
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Li Z, Jakkula L, Hussey RS, Tamulonis JP, Boerma HR (2001) SSR mapping and confirmation of the QTL from PI96354 conditioning soybean resistance to southern root-knot nematode. Theor Appl Genet 103(8):1167–1173. doi:10.1007/s001220100672
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25(15):1966–1967. doi:10.1093/bioinformatics/btp336
Lim KY, Matyasek R, Kovarik A, Leitch AR (2004) Genome evolution in allotetraploid Nicotiana. Biol J Linn Soc 82(4):599–606. doi:10.1111/j.1095-8312.2004.00344.x
Lim KY, Kovarik A, Matyasek R, Chase MW, Clarkson JJ, Grandbastien MA, Leitch AR (2007) Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol 175(4):756–763. doi:10.1111/j.1469-8137.2007.02121.x
Liu D, Ma C, Hong W, Huang L, Liu M, Liu H, Zeng H, Deng D, Xin H, Song J, Xu C, Sun X, Hou X, Wang X, Zheng H (2014) Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS One 9(6):e98855. doi:10.1371/journal.pone.0098855
Lu XP, Gui YJ, Xiao BG, Li YP, Tong ZJ, Liu Y, Bai XF, Wu WR, Xia L, Huttner E, Kilian A, Fan LJ (2013) Development of DArT markers for a linkage map of flue-cured tobacco. Chin Sci Bull 58(6):641–648. doi:10.1007/s11434-012-5453-z
Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17(2):240–248. doi:10.1101/Gr.5681207
Moon HS, Nicholson JS, Heineman A, Lion K, van der Hoeven P, Hayes AJ, Lewis RS (2009a) Changes in genetic diversity of US flue-cured tobacco germplasm over seven decades of cultivar development. Crop Sci 49(2):498–508. doi:10.2135/cropsci2008.05.0253
Moon HS, Nifong JM, Nicholson JS, Heineman A, Lion K, van der Hoeven R, Hayes AJ, Lewis RS (2009b) Microsatellite-based analysis of tobacco (Nicotiana tabacum L.) genetic resources. Crop Sci 49(6):2149–2159. doi:10.2135/cropsci2009.01.0024
Ooijen V (2004) MapQTL® 5, software for the mapping of quantitative trait loci in experimental populations. Kyazma BV, Wageningen
Pfender WF, Saha MC, Johnson EA, Slabaugh MB (2011) Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne. Theor Appl Genet 122(8):1467–1480. doi:10.1007/s00122-011-1546-3
Pilet-Nayel ML, Muehlbauer FJ, McGee RJ, Kraft JM, Baranger A, Coyne CJ (2002) Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor Appl Genet 106(1):28–39. doi:10.1007/s00122-002-0985-2
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One. doi:10.1371/journal.pone.0032253
Potato Genome Sequencing C, Xu X, Pan S et al (2011) Genome sequence and analysis of the tuber crop potato. Nature 475(7355):189–195. doi:10.1038/nature10158
Renny-Byfield S, Chester M, Kovarik A, Le Comber SC, Grandbastien MA, Deloger M, Nichols RA, Macas J, Novak P, Chase MW, Leitch AR (2011) Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol Biol Evol 28(10):2843–2854. doi:10.1093/molbev/msr112
Sierro N, Battey JN, Ouadi S, Bovet L, Goepfert S, Bakaher N, Peitsch MC, Ivanov NV (2013a) Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol 14(6):R60. doi:10.1186/gb-2013-14-6-r60
Sierro N, van Oeveren J, van Eijk MJ, Martin F, Stormo KE, Peitsch MC, Ivanov NV (2013b) Whole genome profiling physical map and ancestral annotation of tobacco Hicks Broadleaf. Plant J 75(5):880–889
Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5:3833. doi:10.1038/ncomms4833
Sun XW, Liu DY, Zhang XF et al (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One. doi:10.1371/journal.pone.0058700
Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB et al (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132(4):1141–1160
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106(3):411–422. doi:10.1007/s00122-002-1031-0
Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485(7400):635–641. doi:10.1038/nature11119
van Os H, Stam P, Visser RGF, van Eck HJ (2005) SMOOTH: a statistical method for successful removal of genotyping errors from high-density genetic linkage data. Theor Appl Genet 112(1):187–194. doi:10.1007/s00122-005-0124-y
Wang X, Bennetzen JL (2015) Current status and prospects for the study of Nicotiana genomics, genetics, and nicotine biosynthesis genes. Mol Genet Genomics 290(1):11–21. doi:10.1007/s00438-015-0989-7
Wang K, Wang Z, Li F et al (2012a) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44(10):1098–1103. doi:10.1038/ng.2371
Wang N, Fang LC, Xin HP, Wang LJ, Li SH (2012b) Construction of a high-density genetic map for grape using next generation restriction-site associated DNA sequencing. BMC Plant Biol 12:148. doi:10.1186/1471-2229-12-148
West MA, van Leeuwen H, Kozik A, Kliebenstein DJ, Doerge RW, St Clair DA, Michelmore RW (2006) High-density haplotyping with microarray-based expression and single feature polymorphism markers in Arabidopsis. Genome Res 16(6):787–795. doi:10.1101/gr.5011206
Wu YH, Bhat PR, Close TJ, Lonardi S (2008) Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. doi:10.1371/journal.pgen.1000212
Wu FN, Eannetta NT, Xu YM, Plieske J, Ganal M, Pozzi C, Bakaher N, Tanksley SD (2010) COSII genetic maps of two diploid Nicotiana species provide a detailed picture of synteny with tomato and insights into chromosome evolution in tetraploid N. tabacum. Theor Appl Genet 120(4):809–827. doi:10.1007/s00122-009-1206-z
Wu J, Wang Z, Shi Z et al (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23(2):396–408. doi:10.1101/gr.144311.112
Yang BC, Xiao BG, Chen XJ, Shi CH (2007) Assessing the genetic diversity of tobacco germplasm using intersimple sequence repeat and inter-retrotransposon amplification polymorphism markers. Ann Appl Biol 150(3):393–401. doi:10.1111/j.1744-7348.2007.00139.x
Zhang HY, Liu XZ, Li TS, Yang YM (2006) Genetic diversity among flue-cured tobacco (Nicotiana tabacum L.) revealed by amplified fragment length polymorphism. Bot Stud 47(3):223–229
Zhang Y, Wang L, Xin H, Li D, Ma C, Ding X, Hong W, Zhang X (2013) Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol 13:141. doi:10.1186/1471-2229-13-141
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single-nucleotide polymorphisms in soybean. Genetics 163(3):1123–1134
Acknowledgments
This work was supported by China National Public Industrial (Agriculture) Specific Project (no. 201203091).
Author information
Authors and Affiliations
Corresponding author
Additional information
Daping Gong and Long Huang contributed equally to this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gong, D., Huang, L., Xu, X. et al. Construction of a high-density SNP genetic map in flue-cured tobacco based on SLAF-seq. Mol Breeding 36, 100 (2016). https://doi.org/10.1007/s11032-016-0514-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0514-7