Abstract
Quality protein maize (QPM; Zea mays L.) has effectively enhanced levels of the amino acids, lysine, and tryptophan, over normal maize and provided balanced dietary protein for the health and development of monogastric animals and humans. However, as in normal maize, QPM varieties are low in provitamin A (ProVA), a precursor of vitamin A, which can lead to vitamin A deficiency in humans when maize is a significant part of their diet. In this study, maize inbred Hp321-1 carrying the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 that can enhance levels of ProVA, was used as donor for improving ProVA in QPM inbred lines CML161 and CML171. Functional markers for identifying the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 were used in foreground selection, and simple sequence repeat markers were used in background selection for the BC1F1, BC2F1, and BC2F2 generations. The background recovery rates were 77.4 and 84.5 % for CML161 and CML171 populations, respectively, in the BC1F1 generation, and 89.9 and 92.1 % in the BC2F2 generation. With foreground and background selection, the mean ProVA concentration has been improved from 1.60 µg g−1 in the parent of CML161 to 5.25 µg g−1 in its BC2F3 offspring, from 1.80 µg g−1 in the parent of CML171 to 8.14 µg g−1 in its BC2F3 offspring while maintaining similar QPM characteristics of the recurrent parents. The success from this study offers maize breeders a procedure for increasing ProVA in QPM lines, which will greatly mitigate vitamin A deficiency and protein-energy malnutrition in developing countries.
Similar content being viewed by others
Introduction
Maize is the staple food and main source of energy for millions of poor people living in developing countries, including China (especially Southwestern China), India, and many countries in Africa and Latin America (IITA 2009; Nuss and Tanumihardjo 2010; Li et al. 2012). As most of the maize varieties do not contain enough provitamin A (ProVA), the precursor that leads to the formation of vitamin A, people heavily relying on maize diet may have health problems, such as malnutrition, specifically vitamin A deficiency (VAD) (IITA 2009). The VAD may retard growth, increase risk of diseases (e.g., macular degeneration), and cause reproductive disorders in humans (West 1991; Aguayo and Baker 2005; West and Darnton-Hill 2008). Fortunately, maize germplasm has tremendous genetic diversity for ProVA content. There are germplasm available with high levels of ProVA carotenoids, including α-, β-carotene and γ-cryptoxanthin (Pixley et al. 2013). Thus, breeding maize for increased levels of ProVA would be an economical and efficient way to address VAD, especially in the developing world (Yan et al. 2010; Zhang et al. 2012).
ProVA usually accounts for only 10–20 % of the total carotenoids in the maize kernel, and most yellow maize hybrids have <2 μg g−1 of ProVA (Ortiz-Monasterio et al. 2007; Pixley et al. 2013). A preliminary target level of 15 μg g−1 for ProVA in maize has been set for the HarvestPlus project. This target level of ProVA has been based on three assumptions: (1) gross daily intake of 400 g for adults and 200 g for children; (2) bioavailability ratio of 12:1 μg (retinol activity equivalent); and (3) 50 % retention after processing (Bouis et al. 2011). Following many years of efforts toward increasing ProVA concentrations in maize hybrids, some elite inbreds and maize breeding stocks with substantially high ProVA levels have been developed by plant breeders via the biofortification method, with some levels reaching 15–20 µg g−1 ProVA (Babu et al. 2013).
ProVA carotenoids in maize kernels may lead to different colors in the endosperm, varying from light yellow to dark orange (Weber 1987). However, there is a low correlation between visual grain color and total carotenoids, β-carotene, and β-cryptoxanthin in diverse inbreds, and screening for high ProVA concentration based on kernel color is not considered reliable (Harjes et al. 2008; Mishra and Singh 2010). Visible light range (400–1100 nm)-based methods, such as near-infrared reflectance spectroscopy (NIRS), have successfully been applied to detect total carotenoid and major carotenoids, such as lutein and zeaxanthin, but not ProVA carotenoid concentrations (Berardo et al. 2004). Another method used for measuring ProVA is high-performance liquid chromatography (HPLC). Though it has been used widely for measuring ProVA concentrations, it is expensive, time-consuming, and has low throughput, limiting its use for routine screening in conventional maize breeding programs (Pfeiffer and McClafferty 2007).
The ProVA carotenoid biosynthesis pathway is well studied in plants, and the key genes controlling critical steps in the pathway have been successfully identified (Hirschberg 2001; DellaPenna and Pogson 2006; Giuliano et al. 2008; Cazzonelli and Pogson 2010; Farré et al. 2010). In maize, five genes play important roles in the final accumulation of ProVA in the grain. One gene encoding phytoene synthase (PSY1) with two favorable alleles is associated with variation in total carotenoids (Fu et al. 2013b). Another gene encoding lycopene epsilon cyclase (lcyE) with four favorable alleles is associated with variation in the ratio of various carotenoids in the α- to β-branches of the carotenoid biosynthesis pathway (Harjes et al. 2008). A third gene, crtRB3, encodes α-carotene hydroxylase enzyme and a fourth gene, ZEP1, controls zeaxanthin epoxidase; both of these genes have been known to play a role in the carotenoid metabolic pathway (Vallabhaneni and Wurtzel 2009; Zhou et al. 2012). A fifth gene encodes β-carotene hydroxylase enzyme (crtRB1) with three favorable alleles and it has a significant impact on variation for β-carotene concentration in the endosperm (Fu et al. 2013a). Yan et al. (2010) found that ProVA concentrations of haplotypes with CrtRB1-5′TE and CrtRB1-3′TE favorable alleles were 5.2-fold higher than those of all other haplotypes. Babu et al. (2013) further reported that CrtRB1 clearly had a much larger effect on ProVA concentration than LcyE. However, limited information is available about applying the marker-assisted backcross (MAB) method to select favorable alleles of the CrtRB1 gene for increasing ProVA concentrations in maize grain.
Marker-assisted selection (MAS) is regarded as a key method for increasing ProVA concentrations in maize (Prasanna et al. 2010). Two genes, CrtRB1 on chromosome 10 (Yan et al. 2010) and LcyE on chromosome 8 (Harjes et al. 2008), have the most significant effect on ProVA concentrations in maize grain (Babu et al. 2013). The effectiveness of molecular marker polymorphisms in linking LcyE and CrtRB1 to ProVA concentrations has been verified using 26 tropical maize populations, and the functional gene markers for high ProVA concentration have been used in MAS (Azmach et al. 2013). As Benchimol et al. (2005) pointed out that through a backcross breeding program, source genes related to high ProVA concentration can be integrated into genotypes with elite agronomic traits of the recurrent parents. However, one of the major limitations is the long period of time required for the backcross procedure. Therefore, molecular markers are important tools for accelerating the recovery of recurrent parent genome as well as assisting in the selection of plants that carry a desired marker linked to high ProVA concentration (Bouchez et al. 2002; Oliveira et al. 2008). Marker-assisted backcrossing is highly suited to monitoring the degree of similarity of the lines to the recurrent parent.
The QPM maize inbred lines CML161 and CML171 have been derived from the CIMMYT G25QPM germplasm and have been widely used in genetic studies and in several breeding programs (Fan et al. 2001; Amiruzzaman et al. 2011; Setter et al. 2011; Fan et al. 2014). The maize commercial hybrids HQ-2000 (Vietnam), INIA (Peru), and Yunyou 78 (China) have been developed utilizing CML161 (Prasanna et al. 2001; Fan et al. 2006); other commercial hybrids including Yunrui 8, Yunyou 89, Qiandan 11, and Quian 2609 (China) utilize CML171 (Cordova 2001; Zhu et al. 2003; Fan et al. 2014). These released hybrids have played a very important role in alleviating the malnutrition of children and have been used as an alternative feed ration for swine and poultry, where conventional sources of lysine are from soybean meal or synthetic lysine (López-Pereira 1992). The integration of the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 for the crtRB1 gene for high ProVA concentration into the widely utilized QPM maize lines CML161 and CML171 would provide a valuable nutritional enhancement to the grain for use in the resource-poor regions of the world. The objectives of this study were: (1) to increase ProVA concentrations of QPM maize lines CML161 and CML171 via MAB using a maize line as donor that carries two favorable alleles of CrtRB1, i.e., crtRB1-5′TE-2 and crtRB1-3′TE-1, for high ProVA concentrations and (2) to evaluate the effectiveness of the MAB procedure for transferring the target genes into local elite maize lines by examining the recovery rates of the recurrent parents.
Materials and methods
Plant materials
The temperate maize inbred line Hp321-1 (provided by Prof. Jianbing Yan, Huazhong Agricultural University) carrying the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 of the crtRB1 gene (ProVA concentration = 9.74 µg g−1), was used as the donor parent (male). Two tropical QPM maize inbred lines, CML161 and CML171, were used as the recurrent female parents. At the lcyE locus, there is no polymorphism among Hp321-1, CML161, and CML171; thus gene pyramiding was not involved.
Foreground selection
Foreground selection (FS) refers to selection of favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1) for higher ProVA concentration. Polymerase chain reaction (PCR) amplification of the functional marker for crtRB1-5′TE-2 was done using the forward primer 5′-TTAGAGCCTCGACCCTCTGTG-3′ and the reverse primer 5′-AATCCCTTTCCATGTTACGC-3′. For crtRB1-3′TE-1, the forward primer was 5′-ACACCACATGGACAAGTTCG-3′ and the reverse primers were 5′-ACACTCTGGCCCATGAACAC-3′ and 5′-ACAGCAATACAGGGGACCAG-3′ (Yan et al. 2010).
Background selection
Background selection (BS) refers to selection of the genetic background of the individuals selected via FS. Two DNA bulks were constructed by mixing equal amounts of DNA from five plants of the donor parent (Hp321-1) and five plants of each recurrent parent CML161 and CML171 for screening parental polymorphic SSR markers. In total, 760 SSRs were retrieved from the Maize Genetics and Genomics Database (http://www.maizegdb.org/) and synthesized by Sangon Biotech (Shanghai, China). A total of 98 polymorphic SSR markers between Hp321-1 and CML161, and 89 polymorphic SSR markers between Hp321-1 and CML171, evenly distributed on 10 maize chromosomes, were used for genotyping the FS-selected individuals of the BC1F1, BC2F1, and BC2F2 populations developed with CML161 and CML171.
Backcross breeding program
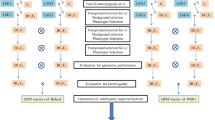
A MAB breeding program was used to integrate the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 from the temperate inbred line Hp321-1 into the tropical QPM maize inbred lines CML161 and CML171, for increasing ProVA concentration. The population development involved two parallel crossing schemes. The QPM inbred line was crossed to Hp321-1 to produce F1, and then backcrossed to CML161 two times to produce BC1F1 and BC2F1. Self-pollination was made to produce BC2F2 and BC2F3 populations. The F1, BC1F1, BC2F1, BC2F2, and BC2F3 populations of CML 171 were produced using the same crossing schemes. In the summer 2010, seeds of CML161, CML171, and Hp321-1 were sown and crosses were made at the YAAS Baiyi Maize Research Station (BMRS, 25°23′N, 102°9′E, 1970 MASL), Kunming, Yunnan, China, to generate F1 populations of CML161 and CML171. In the winter 2010, the F1 was used to develop ‘backcross progenies’ (BC1F1 populations) by backcrossing the F1 with their recurrent parents CML161 and CML171 at the YAAS Gasa Maize Research Station (GMRS, 21°95′N, 100°8′E, 588 MASL), Jinghong, Yunnan, China. In the summer 2011, 597 BC1F1 plants of the CML161 population and 462 BC1F1 plants of the CML171 population were selected via FS and then screened with BS for recovering individuals with ≥80.0 % genetic similarity to the recurrent parent to develop ‘backcross progenies’ BC2F1 populations by backcrossing the BC1F1 with their recurrent parent CML161 and CML171 at the BMRS. In the winter 2011, 779 BC2F1 plants of the CML161 population and 1055 BC2F1 plants of the CML171 population selected via FS were further used to develop ‘selfed progeny’ BC2F2 at the GMRS. In the summer 2012, 1428 BC2F2 plants of the CML161 population and 1554 BC2F2 plants of the CML171 population were planted at the BMRS, and 2452 BC2F2 plants of the CML161 population and 2775 BC2F2 plants of the CML171 population were planted at the Yanshan Maize Research Station (YMRS, 23°60′N, 104°4′E, 1570 MASL), Yanshan, Yunnan, China. Each BC2F2 plant of CML161 population and CML171 population at both locations was selected via FS and then screened for a recovery rate of ≥90.0 % of the recurrent parent via BS, to generate BC2F3 for phenotyping for ProVA concentration, lysine and tryptophan contents. The two parallel crossing schemes for CML161 and Hp321-1, and CML171 and Hp321-1 are shown in Supplementary Fig. 1.
Genotyping
A modified CTAB method (Dellaporta et al. 1983) was used for DNA extraction. The reaction for genotyping with the two functional markers in FS consisted of a total volume of 15 μl containing 30 ng genomic DNA, 0.2 μM of each primer, 1.5 μl of 10× Taq DNA polymerase buffer (20 mM MgCl2), 0.1 mM of each dNTPs (TransGen Biotech, China), and l U of Taq DNA polymerase (TransGen Biotech, China). For parental polymorphic SSR markers in BS, a modified PCR reaction according to McCouch et al. (2002) was used. The reaction consisted of a total volume of 10 μl containing 20 ng genomic DNA, 0.3 μM of each primer, 1.0 μl of 10× Taq DNA polymerase buffer (20 mM MgCl2), 0.1 mM of each dNTPs (TransGen Biotech, China), and l U of Taq DNA polymerase (TransGen Biotech, China).
A modified PCR program (touchdown PCR) was used in profiling functional markers for the favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1) in plants to avoid the amplification of nonspecific, spurious PCR products. The reaction profile with Mastercycler® gradient (Eppendorf, Germany) was performed using the following protocol: First initial hold was at 95 °C for 5 min. The second hold starting with denaturation step at 95 °C for 1 min, annealing at 64 °C for 1 min (10 cycles for crtRB1-5′TE-2, and 19 cycles for crtRB1-3′TE-1, reducing 0.5 °C per cycle), and the extension step at 72 °C for 1 min. The third hold starting at a denaturation temperature of 95 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min with 27 cycles for crtRB1-5′TE-2 and 19 cycles for crtRB1-3′TE-1. The final extension step was done at 72 °C for 10 min. The amplified fragments were resolved in a 2 % agarose gel for analyzing the amplicons.
In profiling of polymorphic SSR markers in BS, the reaction profile was 5 min at 94 °C with 34 cycles of 45 s at 95 °C, and 45 s at 57 °C annealing, and 1 min at 72 °C and 10 min at 72 °C for final extension. The PCR products were separated via electrophoresis on 6 % polyacrylamide gel.
ProVA concentration estimation in parents and the bulked BC2F3 populations
ProVA concentration was estimated according to the modified method of Kimura and Rodrigeuz (2002). A 20-kernel sample was collected from the middle of the ear on each individual plant sampled from the BC2F3 population with the favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1) and from the parental inbreds CML161 and CML171 per se. A total of 127 and 145 ears were sampled from the CML161 and CML171 BC2F3 populations at the BMRS, and 158 and 133 ears were sampled from the respective BC2F3 populations at the YMRS. Five ears were sampled from CML161 and CML171. All samples were mixed and ground with a Cyclotec™ 1093 (FOSS TECATOR) sample mill. A 0.5-g sample of ground grain was placed in the extraction tube, 6 ml of ethanol plus 0.1 % butylated hydroxyl toluene was added, mixed and incubated at 85 °C for 5 min, followed by the addition of 500 μl of 80 % potassium hydroxide (w/v) and incubated at 85 °C for 10 min. After saponification, 3 ml of cold dH2O was added, followed by incubation on ice. A 200 μl internal standard of β-Apo-8′-carotenal and 4 ml hexane were added. The sample was vortexed and centrifuged at 1200×g, and the top hexane layer was transferred to a new tube. The remaining aqueous layer was extracted twice with 3 ml hexane. The combined hexane layers were freeze-dried and stored at −85 °C. Prior to HPLC injection, the carotenoids were reconstituted in 500 μl of acetonitrile:methanol:methylene chloride (45:20:35). All extraction procedures were repeated three times from the ground samples, and each extract was used in separated HPLC analysis.
Carotenoids were separated by HPLC (Agilent-1200) at DL (Shanghai) Company Limited using an YMC CT99S05-2546WT C30 Carotenoid Column. One hundred microliters of the sample was loaded into glass injection vials, and 50 μl was injected at a flow rate of 1.8 ml min−1. The mobile phase consisted of acetonitrile:methanol:methylene chloride at 75:20:4, containing 0.05 % triethylamine (TEA) and 0.1 % BHT. A multi-wavelength detector was set at 450 nm and used to detect the absorbance of carotenoids. ProVA concentration (μg g−1 of dry matter) was calculated as the sum of β-carotene and half of each of β-cryptoxanthin and α-carotene concentrations.
Lysine and tryptophan analysis in parents and the bulked BC2F3 populations
Lysine and tryptophan concentrations were estimated according to the method of Galicia et al. (2008) for a 20-kernel sample from each individual BC2F3 with the favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1), and the parents. After grinding and defatting treatment, 100 mg of the powder for the lysine test was placed in 15 ml extraction tubes, and 3 ml papain solution was added, then mixed and incubated at 64 °C for 16 h. The samples were centrifuged at 2500 rpm for 5 min, and 1 ml of the supernatant was transferred to a new tube and 0.1 ml of 2-chloro-3,5-dinitropyridine reagent (Sigma-Aldrich, America) was added. The mixture was incubated for 2 h at room temperature and then 5 ml of 1.2 N HCl and 5 ml of ethyl acetate were added. The upper phase was removed, and the absorbance was measured by a spectrophotometer (HITACHI U3900/3900H, Japan) at 390 nm, and the lysine concentration was calculated using the following formula:
where A = absorbance at 390 nm.
After the grinding and defatting treatment, 80 mg of the powder for the tryptophan test was placed in a 15-ml extraction tube, and 5 ml of papain solution was added, then mixed, and incubated at 64 °C for 16 h. The samples were centrifuged at 3600×g for 5 min; 1 ml of the supernatant was transferred to a glass tube, and 3 ml of the colorimetric reagent (Sigma-Aldrich, America) was added. The sample was mixed and incubated at 64 °C for 30 min, and the absorbance was measured by a spectrophotometer (HITACHI U3900/3900H, Japan) at 560 nm, and the tryptophan concentration was calculated using the following formula:
where A = absorbance at 560 nm.
Statistical analysis
The gel bands were scored as A for the P1 functional allele (named as genome donor, GD), B for the P2 functional allele (named as genome recurrent, GR), H for the F1 pattern of P1 and P2 functional alleles, and U for an unidentified band. The percentage of the P1 recurrent parent genome present in each genotyped plant was estimated using the expression GR% = [B + (0.5H)/(B + H + A)] × 100 %, corresponding to the number of B functional alleles present in the genotype of each genotyped plant (Benchimol et al. 2005). The introgressed locus was taken from the computation of the total recovery of recurrent parent genotypes. Analysis of variance and calculation of standard deviation were performed using SAS 9.1 software (SAS Institute Inc 2004) for ProVA, lysine, and tryptophan concentration in the BC2F3 individuals, and the recovery rate of the recurrent parent following BS.
Results
Polymorphism of functional markers and SSR markers between parents
The two functional markers of crtRB1 gene were polymorphic between the donor parent (Hp321-1) and the recurrent parents CML161 and CML171 (Supplementary Fig. 2). The donor parent Hp321-1 had the favorable allele crtRB1-5′TE-2 with a 600-bp band, while unfavorable allele crtRB1-5′TE-1 had an 800-bp band corresponding to the recurrent parents CML161 and CML171. Furthermore, the donor parent Hp321-1 carried the favorable allele crtRB1-3′TE-2 with a 543-bp band, while the recurrent parents exhibited the unfavorable allele crtRB1-3′TE-2 with a 296 + 875-bp band in CML161, and crtRB1-3′TE-3 with a 296 + 1221-bp band in CML171. The polymorphic markers were codominant, which rendered them usable for the MAB of the corresponding target genes. Therefore, the functional markers of favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1) were used for FS selection.
In total, 760 SSR markers distributed across the 10 chromosomes of the maize genome were selected for the screening of polymorphisms between the three parent lines. Of these markers, 98 polymorphic SSR markers (12.9 %) between Hp321-1 and CML161, and 89 polymorphic SSR markers (11.7 %) between Hp321-1 and CML171 were identified for BS selection.
Success in transferring the favorable alleles from the donor to recurrent parents
The two recipient parents, CML161 and CML171, were used as recurrent parents to cross to the donor parent, Hp321-1, which had two favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1) for high ProVA concentrations to produce the F1, BC1F1, BC2F1, and BC2F2 generations. In the segregating generations, i.e., BC1F1, BC2F1, and BC2F2, both favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1) were detected (Table 1). The results showed that crtRB1-5′TE-2 and crtRB1-3′TE-1 alleles had linkage in F1 and BC1F1 generations, and no linkage in BC2F2 generation.
In the BC1F1 generation, 597 plants of the CML161 population and 462 plants of the CML171 population were selected and FS was performed for the target alleles (Supplementary Fig. 3). A total of 290 individuals carried the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 in the CML161 population, and 218 individuals in the CML171 population. In the BC2F1 generation, 779 plants of the CML161 and 1055 plants of the CML171 were chosen, and FS was applied for the two target favorable alleles (Supplementary Fig. 4). A total of 410 individuals had the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 in the CML161 population, and 497 individuals in the CML171 population. Chi-square tests showed that segregation ratios for the favorable allele combination of crtRB1-5′TE-2 and crtRB1-3′TE-1 in four populations of BC1F1 and BC2F1 derived from CML161 and CML171 were a good fit to the Mendelian 1:1 ratio expected for one gene-pair inheritance (P > 0.05).
In the BC2F2 generation, 1428 plants of the CML161 population and 1554 plants of the CML171 population were selected at the BMRS and FS was performed for the target favorable alleles (Supplementary Fig. 5). In total, 738 individuals carried the favorable allelic combination of crtRB1-5′TE-2 and crtRB1-3′TE-1 in the CML161 population, and 857 individuals in the CML171 population. At the YMRS, 2452 plants of the CML161 population and 2775 plants of the CML171 population were selected and FS was performed for the favorable alleles. In total, 1129 individuals had the favorable allelic combination of crtRB1-5′TE-2 and crtRB1-3′TE-1 in the CML161 population and 1315 individuals in the CML171 population. Chi-square test showed that segregation ratio for the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 in the BC2F2 population of CML161 was a good fit to the Mendelian 1:2:1 ratio expected for one gene-pair inheritance (P > 0.05), while CML171 population at the BMRS did not fit this ratio (P < 0.05). At the YMRS, the segregation ratios for the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 for both populations (CML161 and CML171) did not fit the Mendelian 1:2:1 ratio (P < 0.05).
The results demonstrated that favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 for high ProVA concentration were successfully transferred into the two QPM maize lines. The two favorable alleles, crtRB1-5′TE-2 and crtRB1-3′TE-1, displayed a 1:1 Mendelian ratio in F1 and BC1F1 generation and segregated in an approximately 1:2:1 ratio in BC2F2 generation.
Improvement in the level of ProVA in kernel of QPM maize lines
In total, the seeds from 127 and 145 plants of BC2F3 generation of CML161 and CML171 populations with the favorable allele combination of crtRB1-5′TE-2 and crtRB1-3′TE-1 at the BMRS, and 158 and 133 plants of CML161 and CML171 populations with the favorable allele combination of crtRB1-5′TE-2 and crtRB1-3′TE-1 at the YMRS were sampled. The ProVA concentrations of the bulked BC2F3 populations were tested using HPLC, and the results are presented in Fig. 1. The mean ProVA concentrations of the BC2F3 generation of CML161 population at the BMRS and YMRS stations were 5.21 and 5.28 µg g−1, with a mean of 5.25 µg g−1, which was significantly higher than that of recurrent parent CML161, with ProVA concentrations of 1.52 and 1.68 µg g−1, with a mean of 1.60 µg g−1, respectively (P < 0.01). Similar results were obtained for the second parallel population of CML171. The mean ProVA concentrations of the BC2F3 generation of CML171 population at the BMRS and YMRS stations were, respectively, 7.58 and 8.70 µg g−1, with a mean of 8.14 µg g−1, which was significantly higher than that of recurrent parent CML171, with the ProVA concentrations of 1.74 and 1.85 µg g−1, with a mean of 1.80 µg g−1, respectively (P < 0.01). The results showed that QPM inbred lines CML161 and CML171 introgressed with the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 had 3.28-fold and 4.52-fold increase in the mean ProVA concentration, which indicated that the MAB procedure was effective in transferring the favorable crtRB1 gene and increasing the ProVA level in maize. Generally, the level of ProVA at the YMRS tended to be higher than that at the BMRS; however, the difference was not significant. The results indicated that ProVA concentrations of maize might be somewhat affected by genotype x environment interaction, but the ProVA was mainly controlled by genetic factors.
Effectiveness of the marker-assisted backcross procedure
Among 760 SSR primers surveyed, 98 polymorphic markers between CML161 and HP321-1, and 89 polymorphic markers between CML171 and HP321-1 were used for selection in each segregating generation, i.e., BC1F1 and BC2F2 (Table 2) and the results presented as genetic background recovery rate in Fig. 2. In the BC1F1 generation, 276 plants from the CML161 population were tested and the results showed that the genetic background recovery rate of selected individuals varied from 63.8 to 90.8 %, with a mean of 77.4 %. This was 3.3 % higher than the mean recovery rate of 74.1 %, varied from 62.7 to 85.5 %, before BS; 210 plants from the CML171 population were analyzed and the results indicated that the genetic background recovery rate of selected individuals ranged from 75.8 to 95.5 %, with a mean of 84.5 %. This was 5.9 % higher than the mean recovery rate of 78.6 %, varied from 67.9 to 84.6 %, before BS. The plants in both populations with a recovery rate of ≥80.0 % were selected for backcrossing.
In the BC2F2 generation, 737 plants from CML161 were tested and the results showed that the genetic background recovery rate of selected individuals with the combination of favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 varied from 85.1 to 94.7 %, with a mean of 89.9 %. This was 7.6 % higher than the mean recovery rate of 82.3 %, varied from 70.1 to 89.2 %, before BS; 702 plants from CML171 were tested and the results indicated that the genetic background recovery rate of selected individuals with the combination of favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 ranged from 88.3 to 96.8 %, with a mean of 92.1 %. This was 5.2 % higher than the mean recovery rate of 86.9 %, varied from 82.4 to 89.5 %, before BS. The results from both populations showed that the individuals of two consecutive backcross generations using molecular MAS had attained similar levels of homozygosity for the favorable alleles of the recurrent parent. The plants carrying favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 in both populations with a recovery rate of ≥90.0 % were selected to generate the BC2F3 generation.
Levels of lysine and tryptophan in the backcross-derived integrated lines
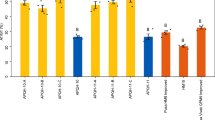
To determine whether the QPM characteristics of the populations were maintained, seeds from BC2F3 generation of CML161 and CML171 populations and from the parental inbreds CML161 and CML171 per se were analyzed for lysine and tryptophan concentration at both locations. The results are presented in Fig. 3. Figure 3 shows that the mean lysine and tryptophan contents are 0.32 and 0.88 % for inbred line CML161, respectively, and the mean lysine and tryptophan contents for CML171 were 0.35 and 0.80 %, respectively, while the mean lysine and tryptophan contents for backcross-derived lines of BC2F3 from CML161 were 0.30 and 0.85 %, respectively, and the mean lysine and tryptophan contents for backcross-derived lines of BC2F3 from CML171 were 0.35 and 0.82 %, respectively. There were almost no difference in the lysine and tryptophan contents between the BC2F3 lines and the original CML161 and CML171 inbred lines. The results suggested that MAS backcross procedure had improved ProVA levels without decreasing lysine and tryptophan levels in QPM maize lines.
Discussion
The ProVA concentrations of both populations derived from inbred lines CML161 and CML171 at the YMRS were higher than those at the BMRS, while the difference between the two stations was not significant. The enhanced protein quality for the derived populations was also maintained in the backcross-derived lines. The QPM lines with improved ProVA concentration are very important breeding materials for developing QPM maize hybrids with high ProVA for people dependent on maize as a major component of their diet. Therefore, the development and production of QPM hybrids with high ProVA concentrations are an efficient way to deliver biofortified maize for improved health and development benefits to the poor people living in developing countries, such as China (especially Southwestern China), India, and several countries in Africa and Latin America.
Previous investigations have revealed that the differences in expression levels of the two loci LcyE and CrtRB1 were mainly expressed in the endosperm and were not different in embryos (Harjes et al. 2008; Yan et al. 2010). Thus, selecting for favorable mutant alleles of LcyE and/or CrtRB1 could cause undesirable effects in the endosperm. In the present study, MAB technology was used to produce two QPM populations with higher levels of ProVA than those of their parents. In the CML161 and CML171 populations, the mean ProVA levels were up to 5.25 and 8.14 µg g−1, respectively, which were significantly higher than those of recurrent parents CML161 (1.60 µg g−1) and CML171 (1.80 µg g−1) (P = 0.01). Meanwhile, the lines maintained their protein quality with a lysine content of 0.30 and 0.35 %, and a tryptophan content of 0.85 and 0.82 % in CML161 and CML171 populations with the ProVA being 3.28-fold and 4.52-fold higher than those of the QPM parents CML161 and CML171, respectively. Though the target concentration of >15 µg g−1 for ProVA was not achieved, these lines with enhanced levels of ProVA over their original QPM parent lines could be used as parents to develop maize cultivars that meet the dietary needs of both humans and livestock. These improved QPM lines can be used for developing nutritionally enhanced hybrids. This study is the first report for combining functional marker selection for CrtRB1-5′TE and CrtRB1-3′TE favorable alleles with the backcross method to obtain improved ProVA lines while maintaining the lysine and tryptophan level of QPM. The authors hope this effective procedure will be applied by more breeders toward mitigating both VAD and protein-energy malnutrition in developing countries.
The MAS for the CrtRB1 locus appeared to be a reliable strategy for rapidly achieving genetic gains for high ProVA concentration in previous research (Azmach et al. 2013; Babu et al. 2013). However in this study, both microsatellites and MAB selection were used simultaneously. The functional markers of CrtRB1 locus were used to identify the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 for a rapid tripling of ProVA concentration. Meanwhile, when SSR markers were combined for the BS, more than 90 % of the recurrent parent could be recovered via only two backcrosses, while at least three backcrosses would be required in conventional breeding process (Chen et al. 2010). The results indicated that the MAB procedure was an effective strategy for transferring the target genes into local elite maize lines by examining the recovery rates of the recurrent parents. Therefore, this study provided a good example to increase the efficiency in the breeding program for high ProVA level via the BS with SSR markers.
When the functional markers of crtRB1-3′TE alleles were used to screen the progenies, we found that the upper bands for the crtRB1-3′TE-2/3 favorable allele with 845- and 1221-bp were not always present. For example, very few progeny in the BC1F1 generation had the 845- and 1221-bp bands for the crtRB1-3′TE-2/3 favorable allele, while the two bands were frequently displayed in the BC2F2 generation. Compared with a previous report (Yan et al. 2010), the bands of 845- and 1221-bp were much fainter than the bands of 296- and 543-bp. The possible reason might be that the bands of 845- and 1221-bp required more stringent conditions for the PCR reaction. A similar problem was observed in another study (Babu et al. 2013). Therefore, we recommend that the favorable allele of crtRB1-3′TE gene should be identified using high-fidelity DNA polymerase and the progenies can be tested by universal DNA polymerase.
In the FS for the favorable alleles crtRB1-5′TE-2 and crtRB1-3′TE-1 in various segregating generations, we found that at the YMRS, the segregation ratio of BC2F2 generation for CML161 did not fit the Mendelian 1:2:1 ratio expected for one gene inheritance (P < 0.05). Similar results were observed in BC2F2 generation of CML171 at the BMRS and YMRS. A possible reason for this deviation might be that the progenies were artificially selected to have both favorable alleles (crtRB1-5′TE-2 and crtRB1-3′TE-1) or it could be because crtRB1-5′TE-2 and crtRB1-3′TE-1 occurred in a known segregation distortion region (Lu et al. 2002; Babu et al. 2013). We suggest that a larger population size would be required to resolve segregation distortion effects on selection in the breeding program for high ProVA level via functional markers.
References
Aguayo VM, Baker SK (2005) Vitamin A deficiency and child survival in sub-Saharan Africa: a reappraisal of challenges and opportunities. Food Nutr Bull 26:348–355
Amiruzzaman M, Islam MA, Pixley KV, Rohman MM (2011) Heterosis and combining ability of CIMMYT’s tropical × subtropical quality protein maize germplasm. Int J Agric Sustain 3:76–81
Azmach G, Gedil M, Menkir A, Spillane C (2013) Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Biol 13:227
Babu R, Rojas NP, Gao S, Yan J, Pixley K (2013) Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor Appl Genet 126:389–399
Benchimol LL, Souza CL Jr, Souza AP (2005) Microsatellite-assisted backcross selection in maize. Genet Mol Biol 28:789–797
Berardo N, Brenna OV, Amato A, Valoti P, Pisacane V, Motto M (2004) Carotenoids concentration among maize genotypes measured by near infrared reflectance spectroscopy (NIRS). Innov Food Sci Emerg Technol 5:393–398
Bouchez A, Hospital F, Causse M, Gallais A, Charcosset A (2002) Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics 162:1945–1959
Bouis HE, Hotz C, McClafferty B, Meenakshi JV, Pfeiffer WH (2011) Biofortification: a new tool to reduce micronutrient malnutrition. Food Nutr Bull 32:S31–S40
Cazzonelli CI, Pogson BJ (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15:266–274
Chen HM, Zhang YD, Chen W, Kang MS, Tan J, Wang YF, Yang JY, Fan XM (2010) Improving grain yield and yield components via backcross procedure. Maydica 55:145–153
Cordova H (2001) Quality protein maize: improved nutrition and livelihoods for the poor. In: CIMMYT (eds) Maize research highlights 1999–2000. CIMMYT, Mexico, p 26
DellaPenna D, Pogson BJ (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57:711–738
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini-preparation: version II. Plant Mol Biol Rep 1:19–21
Fan XM, Tan J, Huang BH, Liu F (2001) Analyses of combining ability and heterotic group of yellow grain quality protein maize inbreeds. Hereditas 23:547–552
Fan XM, Chen HM, Tan J, Huang YX, Duan ZL, Liu F, Han XR (2006) Analysis of yield combining ability of subtropical tropical quality protein maize inbred line and temperate normal maize inbred line. J Maize Sci 14:12–15 (in Chinese with English abstract)
Fan XM, Zhang YD, Yao WH, Bi YQ, Liu L, Chen HM, Kang MS (2014) Reciprocal diallel crosses impact combining ability, variance estimation, and heterotic group classification. Crop Sci 54:89–97
Farré G, Sanahuja G, Naqvi S, Bai C, Capell T (2010) Travel advice on the road to carotenoids in plants. Plant Sci 179:28–48
Fu JJ, Cheng YB, Linghu J, Yang XH, Kang L, Zhang ZX, Zhang J, He C, Du XM, Peng ZY, Wang B, Zhai LH, Dai CM, Xu JB, Wang WD, Li XR, Zheng J, Chen L, Luo LH, Liu JJ, Qian XJ, Yan JB, Wang J, Wang GY (2013a) RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun 4:2832
Fu ZY, Chai YC, Zhou Y, Yang XH, Warburton ML, Xu ST, Cai Y, Zhang DL, Li JS, Yan JB (2013b) Natural variation in the sequence of PSY1 and frequency of favorable polymorphisms among tropical and temperate maize germplasm. Theor Appl Genet 126:923–935
Galicia L, Nurit E, Rosales A, Palacios B, Rojas N (2008) Maize nutrition quality and plant tissue analysis laboratory. CIMMYT, Mexico, pp 10–17
Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA (2008) Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 26:139–145
Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, Yan J, Buckler ES (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319:330–333
Hirschberg J (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4:210–218
International Institute of Tropical Agriculture (IITA) (2009) Maize production, consumption and harvesting. Retrieved from: www.iita.org (Accessed on: 9 Sep 2011)
Kimura M, Rodrigeuz AD (2002) A scheme for obtaining standards and HPLC quantification of leafy vegetable carotenoids. Food Chem 78:389–398
Li J, Lammerts van Bueren ET, Jiggins J, Leeuwis C (2012) Farmers’ adoption of maize (Zea mays L.) hybrids and the persistence of landraces in Southwest China: implications for policy and breeding. Genet Resour Crop Evol 59:1147–1160
López-Pereira MA (1992) The economics of quality protein maize as an animal feed: case studies of Brazil and El Salvador. CIMMYT, Mexico
Lu H, Romero-Severson J, Bernardo R (2002) Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet 105:622–628
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–220
Mishra P, Singh NK (2010) Spectrophotometry and TLC based characterization of kernel carotenoids in short duration maize. Maydica 55:95–100
Nuss ET, Tanumihardjo SA (2010) Maize: a paramount staple crop in the context of global nutrition. Compr Rev Food Sci Food Saf 9:417–436
Oliveira LK, Melo LC, Brondani C, Peloso MJD, Brondani RPV (2008) Backcross assisted by microsatellite markers in common bean. Genet Mol Res 7:1000–1010
Ortiz-Monasterio JI, Palacios-Rojas N, Meng E, Pixley K, Trethowan R, Peña RJ (2007) Enhancing the mineral and vitamin content of wheat and maize through plant breeding. J Cereal Sci 46:293–307
Pfeiffer WH, McClafferty B (2007) Harvestplus: breeding crops for better nutrition. Crop Sci 47:S88–S105
Pixley K, Rojas NN, Babu R, Mutale R, Surles R, Simpungwe E (2013) Biofortification of maize with provitamin A carotenoids. In: Tanumihardjo SA (ed) Carotenoids and human health SE-17. Humana Press, Totowa, pp 271–292
Prasanna BM, Vasal SK, Kassahun B, Singh NN (2001) Quality protein maize. Curr Sci 81:1308–1319
Prasanna BM, Pixley K, Warburton ML, Xie CX (2010) Molecular marker-assisted breeding options for maize improvement in Asia. Mol Breed 26:339–356
SAS Institute Inc (2004) SAS/STAT user’s guide. SAS Institute Inc, Cary
Setter TL, Yan J, Warburton M, Ribaut JM, Xu Y, Sawkins M, Buckler ES, Zhang Z, Gore MA (2011) Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought. J Exp Bot 62:701–716
Vallabhaneni R, Wurtzel ET (2009) Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol 150:562–572
Weber E (1987) Carotenoids and tocols of corn grain determined by HPLC. J Am Oil Chem Soc 64:1129–1134
West KP (1991) Dietary vitamin A deficiency: effects on growth, infection, and mortality. Food Nutr Bull 13:119–131
West KP, Darnton-Hill I (2008) Vitamin A deficiency. In: Semba RD, Bloem MW (eds) Nutrition and health in developing countries, 2nd edn. Humana Press, Totowa
Yan JB, Kandianis CB, Harjes CE, Bai L, Kim EH, Yang XH, Skinner DJ, Fu ZY, Mitchell S, Li Q, Salas Fernandez MG, Zaharieva M, Babu R, Fu Y, Palacios Rojas N, Li JS, Dellapenna D, Brutnell T, Buckler ES, Warburton ML, Rocheford T (2010) Rare genetic variation at Zea mays crtRB1 increases b-carotene in maize grain. Nat Genet 42:322–327
Zhang X, Pfeiffer WH, Palacios-Rojas N, Babu R, Bouis H, Wang J (2012) Probability of success of breeding strategies for improving pro-vitamin A content in maize. Theor Appl Genet 125:235–246
Zhou Y, Han YJ, Li ZG, Fu Y, Fu ZY, Xu S, Li J, Yan J, Yang X (2012) ZmcrtRB3 encodes a carotenoid hydroxylase that affects the accumulation of α-carotene in maize kernel. J Integr Plant Biol 54:260–269
Zhu YF, Chen ZH, Gao X (2003) The application of exotic maize germplasm in Guizhou. Seeds 4:67–68 (in Chinese with English abstract)
Acknowledgments
The study was supported by the Yunnan Leading Talent Project (2014HA002), National Natural Science Foundation of China Project (31060192), China 948 Project (2015-Z73), and by the Middle-age and Young Academic Leader and Cultivating Talent of Science Foundation of Yunnan Province (2011CI037).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
General scheme for the development of backcross and selfed progenies. The temperate maize inbred line Hp321-1 with favorable alleles crtRB1-5’TE-2 and crtRB1-3’TE-1 for high ProVA concentrations was used as male and donor parent. The tropical QPM maize inbred lines CML161 and CML171 were used as female and recurrent parents. The numbers 1, 2, 3, 4, and 5 refer two populations developed for CML161 and CML171, respectively. (TIFF 121 kb)
Supplementary Fig. 2
Electrophoresis pattern of functional markers between parents for target alleles. (A) Patterns of crtRB1-5’TE-2 allele; (B) Patterns of crtRB1-3’TE-1 allele; 1, DNA marker; 2, Hp321-272; 3, Hp321-3; 4, Hp321-1; 5, Sc55; 6, A619; 7, By804; 8, CML161; 9, CML171; 10, P138. Lanes 2–7 are maize inbred lines with higher ProVA concentration and favorable LcyE and crtRB1 allelic combination. Lanes 8–10 are maize inbred lines with lower ProVA concentration and unfavorable LcyE and crtRB1 allelic combination. (TIFF 195 kb)
Supplementary Fig. 3
Detection of the favorable alleles crtRB1-5’TE-2 and crtRB1-3’TE-1 in BC1F1 generation. (A) Patterns of crtRB1-5’TE-2 allele in the CML161 population, (B) Patterns of crtRB1-3’TE-1 allele in the CML161 population, (C) Patterns of crtRB1-5’TE-2 allele in the CML171 population, (D) Patterns of crtRB1-3’TE-1 allele in the CML171 population. (TIFF 419 kb)
Supplementary Fig. 4
Detection of the favorable alleles crtRB1-5’TE-2 and crtRB1-3’TE-1 in the BC2F1 generation. (A) Patterns of crtRB1-5’TE-2 allele in the CML161 population, (B) Patterns of crtRB1-3’TE-1 allele in the CML161 population, (C) Patterns of crtRB1-5’TE-2 allele in the CML171 population, (D) Patterns of crtRB1-3’TE-1 allele in the CML171 population. (TIFF 392 kb)
Supplementary Fig. 5
Detection of the favorable alleles crtRB1-5’TE-2 and crtRB1-3’TE-1 in the BC2F2 generation. (A) Patterns of crtRB1-5’TE-2 allele in the CML161 population, (B) Patterns of crtRB1-3’TE-1 allele in the CML161 population, (C) Patterns of crtRB1-5’TE-2 allele in the CML171 population, (D) Patterns of crtRB1-3’TE-1 allele in the CML171 population. (TIFF 284 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liu, L., Jeffers, D., Zhang, Y. et al. Introgression of the crtRB1 gene into quality protein maize inbred lines using molecular markers. Mol Breeding 35, 154 (2015). https://doi.org/10.1007/s11032-015-0349-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0349-7