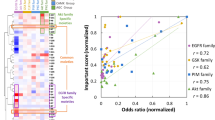
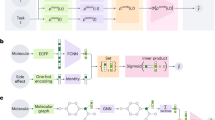
Abstract
Kinase plays a significant role in various disease signaling pathways. Due to the highly conserved sequence of kinase family members, understanding the selectivity profile of kinase inhibitors remains a priority for drug discovery. Previous methods for kinase selectivity identification use biochemical assays, which are very useful but limited by the protein available. The lack of kinase selectivity can exert benefits but also can cause adverse effects. With the explosion of the dataset for kinase activities, current computational methods can achieve accuracy for large-scale selectivity predictions. Here, we present a multimodal multi-task deep neural network model for kinase selectivity prediction by calculating the fingerprint and physiochemical descriptors. With the multimodal inputs of structure and physiochemical properties information, the multi-task framework could accurately predict the kinome map for selectivity analysis. The proposed model displays better performance for kinase–target prediction based on system evaluations.
Graphic abstract







Similar content being viewed by others
Abbreviations
- DTI:
-
Drug–target interactions
- kNN:
-
K-nearest neighbor algorithm
- MT-DNN:
-
Multi-task deep neural network
- DNN:
-
Deep neural network
- ReLU:
-
Rectified Linear Unit
- MST-DNN:
-
Multimodal single-task deep neural network
- QSAR:
-
Quantitative structure–activity relationship
- MCC:
-
Matthews correlation coefficient
- PCA:
-
Principal component analysis
- CNN:
-
Convolutional neural network
References
Ardito F, Giuliani M, Perrone D, Troiano G, Lo Muzio L (2017) The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int J Mol Med 402:271–280. https://doi.org/10.3892/ijmm.2017.3036
Zimmermann J (2009) Interview with Jürg Zimmermann, global head of oncology & exploratory chemistry at Novartis. Future Med Chem 18:1395–1398. https://doi.org/10.4155/Fmc.09.115
Attwood MM, Fabbro D, Sokolov AV, Knapp S, Schiöth HB (2021) Trends in kinase drug discovery: targets, indications and inhibitor design. Nat Rev Drug Discov 2011:839–861. https://doi.org/10.1038/s41573-021-00252-y
Dagher R, Cohen M, Williams G, Rothmann M, Gobburu J, Robbie G, Rahman A, Chen G, Staten A, Griebel D (2002) Approval summary: imatinib mesylate in the treatment of metastatic and/or unresectable malignant gastrointestinal stromal tumors. Clin Cancer Res 810:3034–3038
Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P (2008) Drug target identification using side-effect similarity. Science 3215886:263–266. https://doi.org/10.1126/science.1158140
Peska L, Buza K, Koller J (2017) Drug-target interaction prediction: a Bayesian ranking approach. Comput Methods Programs Biomed 152:15–21. https://doi.org/10.1016/j.cmpb.2017.09.003
Yamanishi Y, Araki M, Gutteridge A, Honda W, Kanehisa M (2008) Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 2413:i232–i240. https://doi.org/10.1093/bioinformatics/btn162
Chen ZH, You ZH, Guo ZH, Yi HC, Luo GX, Wang YB (2020) Prediction of drug-target interactions from multi-molecular network based on deep walk embedding model. Front Bioeng Biotechnol 8:338. https://doi.org/10.3389/fbioe.2020.00338
Wen M, Zhang Z, Niu S, Sha H, Yang R, Yun Y, Lu H (2017) Deep-learning-based drug–target interaction prediction. J Proteome Res 164:1401–1409. https://doi.org/10.1021/acs.jproteome.6b00618
Chen R, Liu X, Jin S, Lin J, Liu J (2018) Machine learning for drug-target interaction prediction. Molecules 239:2208. https://doi.org/10.3390/molecules23092208
Rifaioglu AS, Nalbat E, Atalay V, Martin MJ, Cetin-Atalay R, Doğan T (2020) DEEPScreen: high performance drug–target interaction prediction with convolutional neural networks using 2-D structural compound representations. Chem Sci 119:2531–2557. https://doi.org/10.1039/c9sc03414e
Wan F, Zhu Y, Hu H, Dai A, Cai X, Chen L, Gong H, Xia T, Yang D, Wang M-W (2019) DeepCPI: a deep learning-based framework for large-scale in silico drug screening. Genomics Proteomics Bioinf 175:478–495. https://doi.org/10.1016/j.gpb.2019.04.003
Madhukar NS, Khade PK, Huang L, Gayvert K, Galletti G, Stogniew M, Allen JE, Giannakakou P, Elemento O (2019) A Bayesian machine learning approach for drug target identification using diverse data types. Nat Commun 101:5221. https://doi.org/10.1038/s41467-019-12928-6
Mayr A, Klambauer G, Unterthiner T, Steijaert M, Wegner JK, Ceulemans H, Clevert D-A, Hochreiter S (2018) Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem Sci 924:5441–5451. https://doi.org/10.1039/c8sc00148k
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830. https://doi.org/10.48550/arXiv.1201.0490
Bajorath J (2001) Selected concepts and investigations in compound classification, molecular descriptor analysis, and virtual screening. J Chem Inf Comput Sci 412:233–245. https://doi.org/10.1021/ci0001482
Gao K, Nguyen DD, Sresht V, Mathiowetz AM, Tu M, Wei GW (2020) Are 2D fingerprints still valuable for drug discovery? Phys Chem Chem Phys 2216:8373–8390. https://doi.org/10.1039/d0cp00305k
Thung KH, Wee CY (2018) A brief review on multi-task learning. Multimedia Tools Appl 7722:29705–29725. https://doi.org/10.1007/s11042-018-6463-x
Wenzel J, Matter H, Schmidt F (2019) Predictive multitask deep neural network models for ADME-Tox properties: learning from large data sets. J Chem Inf Model 593:1253–1268. https://doi.org/10.1021/acs.jcim.8b00785
Xu Y, Ma J, Liaw A, Sheridan RP, Svetnik V (2017) Demystifying multitask deep neural networks for quantitative structure–activity relationships. J Chem Inf Model 5710:2490–2504. https://doi.org/10.1021/acs.jcim.7b00087
Chollet F (2018) Keras: The python deep learning library. Astrophysics Source Code Library: ascl 1806:022
Cover T, Hart P (1967) Nearest neighbor pattern classification. IEEE Trans Inf Theory 131:21–27. https://doi.org/10.1109/TIT.1967.1053964
Korotcov A, Tkachenko V, Russo DP, Ekins S (2017) Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol Pharm 1412:4462–4475. https://doi.org/10.1021/acs.molpharmaceut.7b00578
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado G S, Davis A, Dean J, Devin M (2016) Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint https://doi.org/10.48550/arXiv.1603.04467
Klopman G, Kalos AN (1985) Causality in structure—activity studies. J Comput Chem 65:492–506. https://doi.org/10.1002/jcc.540060520
Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 2911:1046–1051. https://doi.org/10.1038/nbt.1990
Li X, Li Z, Wu X, Xiong Z, Yang T, Fu Z, Liu X, Tan X, Zhong F, Wan X (2019) Deep learning enhancing kinome-wide polypharmacology profiling: model construction and experiment validation. J Med Chem 6316:8723–8737. https://doi.org/10.1021/acs.jmedchem.9b00855
Zhao J, Zhang D, Zhang W, Stashko MA, DeRyckere D, Vasileiadi E, Parker RE, Hunter D, Liu Q, Zhang Y (2018) Highly selective MERTK inhibitors achieved by a single methyl group. J Med Chem 6122:10242–10254. https://doi.org/10.1021/acs.jmedchem.8b01229
Sather S, Kenyon KD, Lefkowitz JB, Liang X, Varnum BC, Henson PM, Graham DK (2007) A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood 1093:1026–1033. https://doi.org/10.1182/blood-2006-05-021634
Hua Y, Fang X, Xing G, Xu Y, Liang L, Deng C, Dai X, Liu H, Lu T, Zhang Y (2022) Effective reaction-based de novo strategy for kinase targets: a case study on MERTK inhibitors. J Chem Inf Model 627:1654–1668. https://doi.org/10.1021/acs.jcim.2c00068
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (Grant Nos. 81973182, 82073704, 81803370) State Key Laboratory Innovation Research and Cultivation Fund (Grant No. SKLNMZZCX201812), and “Double World-classes” Construction Program of China Pharmaceutical University (Grant No. CPU2018GF02).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hua, Y., Luo, L., Qiu, H. et al. Multimodal multi-task deep neural network framework for kinase–target prediction. Mol Divers 27, 2491–2503 (2023). https://doi.org/10.1007/s11030-022-10565-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10565-8