Abstract
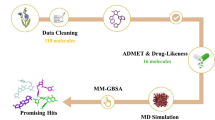
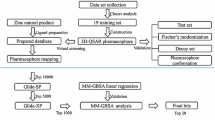
One of the most common malignancies diagnosed and the leading cause of death for cancer-stricken women globally is breast cancer. The molecular subtype affects therapy options because it is a complex disorder with multiple subtypes. By concentrating on receptor activation, mTOR (mammalian target of rapamycin) can be employed as a therapeutic target. The goal of this work was to screen a number of inhibitors produced from Hibiscus rosa-sinensis for possible target to inhibit the mTOR and to determine which has the greatest affinity for the receptor. Primarily, the ionization states of the chosen compounds were predicted using the ChemAxon web platform, and their pKa values were estimated. Given the significance of interactions between proteins in the development of drugs, structure-based virtual screening was done using AutoDock Vina. Approximately 120 Hibiscus components and ten approved anti-cancer drugs, including the mTOR inhibitor everolimus, were used in the comparative analysis. By using Lipinski's rule of five to the chosen compounds, the ADMET profile and drug-likeness characteristics were further examined to assess the anti-breast cancer activity. The compounds with the highest ranked binding poses were loaded using the SeeSAR tool and the HYDE scoring to give interactive, desolvation, and visual ΔG estimation for ligand binding affinity assessment. Following, the prospective candidates underwent three replicas of 100 ns long molecular dynamics simulations, preceded with MM-GBSA binding free energy calculation. The stability of the protein–ligand complex was determined using root mean square deviation (RMSD), root mean square fluctuation (RMSF), and protein–ligand interactions. The results demonstrated that the best mTOR binding affinities were found for stigmastadienol (107), lupeol (66), and taraxasterol acetate (111), which all performed well in comparison to the control compounds. Thus, bioactive compounds isolated from Hibiscus rosa-sinensis could serve as lead molecules for the creation of potent and effective mTOR inhibitors for the breast cancer therapy.
Graphical abstract













Similar content being viewed by others
Data availability
All data generated during this study are included in supplementary information files.
Code availability
Not applicable.
References
Wilkinson L, Gathani T (2022) Understanding breast cancer as a global health concern. Br J Radiol 95:20211033
Dai X, Xiang L, Li T, Bai Z (2016) Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J Cancer 7:1281
Chan S, Scheulen ME, Johnston S, Mross K, Cardoso F, Dittrich C, Eiermann W, Hess D, Morant R, Semiglazov V (2005) Phase II study of temsirolimus (CCI-779), a novel inhibitor of mTOR, in heavily pretreated patients with locally advanced or metastatic breast cancer. J Clin Oncol 23:5314–5322. https://doi.org/10.1200/JCO.2005.66.130
M. Castellanos, C. Gubern, E. Kadar, mTOR: Exploring a New Potential Therapeutic Target for Stroke, in: Mol. to Med. with MTOR, Elsevier, 2016: pp. 105–122. https://doi.org/10.1016/B978-0-12-802733-2.00012-8.
Wong M (2013) Mammalian target of rapamycin (mTOR) pathways in neurological diseases. Biomed J 36:1
Tekirdag KA, Ozturk DG, Gozuacik D (2015) Regulation of Autophagy by microRNAs. In: Cancer A, Pathol O (eds) Inflammation, Immunity, Infect. Elsevier, Aging, pp 81–101
Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149:274–293. https://doi.org/10.1016/j.cell.2012.03.017
Zou Z, Tao T, Li H, Zhu X (2020) mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci 10:1–11. https://doi.org/10.1186/s13578-020-00396-1
Unni N, Arteaga CL (2019) Is dual mTORC1 and mTORC2 therapeutic blockade clinically feasible in cancer? JAMA Oncol 5:1564–1565. https://doi.org/10.1001/jamaoncol.2019.2525
Peng Y, Wang Y, Zhou C, Mei W, Zeng C (2022) PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front Oncol 12:819128
Engelman JA, Luo J, Cantley LC (2006) The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7:606–619. https://doi.org/10.1038/nrg1879
Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science. https://doi.org/10.1126/science.1096502
Gera J, Lichtenstein A (2011) The mammalian target of rapamycin pathway as a therapeutic target in multiple myeloma. Leuk Lymphoma 52:1857–1866. https://doi.org/10.3109/10428194.2011.580478
Dowling RJO, Topisirovic I, Fonseca BD, Sonenberg N (2010) Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim Biophys Acta 1804:433–439. https://doi.org/10.1016/j.bbapap.2009.12.001
Vittorio S, Gitto R, Adornato I, Russo E, De Luca L (2021) In silico strategy for targeting the mTOR kinase at rapamycin binding site by small molecules. Molecules 26:1103. https://doi.org/10.3390/molecules26041103
Zulkipli NN, Zakaria R, Long I, Abdullah SF, Muhammad EF, Wahab HA, Sasongko TH (2020) In silico analyses and cytotoxicity study of asiaticoside and asiatic acid from malaysian plant as potential mTOR inhibitors. Molecules 25:3991. https://doi.org/10.3390/molecules25173991
Zoncu R, Efeyan A, Sabatini DM (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12:21–35. https://doi.org/10.1038/nrm3025
Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168:960–976. https://doi.org/10.1016/j.cell.2017.02.004
Mossmann D, Park S, Hall MN (2018) mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer 18:744–757. https://doi.org/10.1038/s41568-018-0074-8
Hare SH, Harvey AJ (2017) mTOR function and therapeutic targeting in breast cancer. Am J Cancer Res 7:383
Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y (2019) Targeting mTOR for cancer therapy. J Hematol Oncol 12:1–19. https://doi.org/10.1186/s13045-019-0754-1
Adhirajan N, Kumar TR, Shanmugasundaram N, Babu M (2003) In vivo and in vitro evaluation of hair growth potential of Hibiscus rosa-sinensis Linn. J Ethnopharmacol 88:235–239
Faraji MH, Tarkhani AHH (1999) The effect of sour tea (Hibiscus sabdariffa) on essential hypertension. J Ethnopharmacol 65:231–236
P.C. Ochani, P. D’Mello, Antioxidant and antihyperlipidemic activity of Hibiscus sabdariffa Linn. leaves and calyces extracts in rats (2009).
Chiu C-T, Chen J-H, Chou F-P, Lin H-H (2015) Hibiscus sabdariffa leaf extract inhibits human prostate cancer cell invasion via down-regulation of Akt/NF-κB/MMP-9 pathway. Nutrients 7:5065–5087
Tseng T-H, Kao T-W, Chu C-Y, Chou F-P, Lin W-L, Wang C-J (2000) Induction of apoptosis by hibiscus protocatechuic acid in human leukemia cells via reduction of retinoblastoma (RB) phosphorylation and Bcl-2 expression. Biochem Pharmacol 60:307–315
Lin H-H, Chen J-H, Kuo W-H, Wang C-J (2007) Chemopreventive properties of Hibiscus sabdariffa L. on human gastric carcinoma cells through apoptosis induction and JNK/p38 MAPK signaling activation. Chem Biol Interact 165:59–75
Malacrida A, Maggioni D, Cassetti A, Nicolini G, Cavaletti G, Miloso M (2016) Antitumoral effect of Hibiscus sabdariffa on human squamous cell carcinoma and multiple myeloma cells. Nutr Cancer 68:1161–1170
Nguyen C, Baskaran K, Pupulin A, Ruvinov I, Zaitoon O, Grewal S, Scaria B, Mehaidli A, Vegh C, Pandey S (2019) Hibiscus flower extract selectively induces apoptosis in breast cancer cells and positively interacts with common chemotherapeutics. BMC Complement Altern Med 19:1–14
Hsu R-J, Hsu Y-C, Chen S-P, Fu C-L, Yu J-C, Chang F-W, Chen Y-H, Liu J-M, Ho J-Y, Yu C-P (2015) The triterpenoids of Hibiscus syriacus induce apoptosis and inhibit cell migration in breast cancer cells. BMC Complement Altern Med 15:1–9
Vijayakumar S, Yabesh JEM, Arulmozhi P, Praseetha PK (2018) Identification and isolation of antimicrobial compounds from the flower extract of Hibiscus rosa-sinensis L.: in silico and in vitro approaches. Microb Pathog 123:527–535
J.Y. Salib, Polyphenolic Compounds from Flowers of Hibiscus: Characterization and Bioactivity, in: Polyphenols in Plants, Elsevier, 2014: pp. 231–239.
Ateya A-M, El Sayed ZI, Fekry M (2012) Chemical constituents, cytotoxicity, anti-oxidant, hypoglycemic and antihypertensive activities of Egyptian Hibiscus trionum. Aust J Basic Appl Sci 6:756–766
J.F. Morton, Renewed interest in Roselle (Hibiscus sabdariffa l.), the long-forgotten" florida cranberry", in: Proc. Florida State Hortic. Soc., 1974: pp. 415–425.
I.A. Ross, Hibiscus sabdariffa, in: Med. Plants World, Springer, 2003: pp. 267–275.
Ngan LTM, Tan MT, Hoang NVM, Thanh DT, Linh NTT, Hoa TTH, Nuong NTM, Hieu TT (2021) Antibacterial activity of Hibiscus rosa-sinensis L. red flower against antibiotic-resistant strains of Helicobacter pylori and identification of the flower constituents. Brazilian J. Med. Biol. Res. 54:1
Salib JY, Daniel EN, Hifnawy MS, Azzam SM, Shaheed IB, Abdel-Latif SM (2011) Polyphenolic compounds from flowers of Hibiscus rosa-sinensis Linn. and their inhibitory effect on alkaline phosphatase enzyme activity in vitro. Zeitschrift Für Naturforsch C. 66:453–459
Gandhi SP, Lokhande KB, Swamy VK, Nanda RK, Chitlange SS (2019) Computational data of phytoconstituents from Hibiscus rosa-sinensis on various anti-obesity targets. Data Br 24:103994
Salama RB, Ibrahim SA (1979) Ergosterol in Hibiscus sabdariffa seed oil. Planta Med 36:221–222
B.H. Ali, N. Al Wabel, G. Blunden, Phytochemical, pharmacological and toxicological aspects of Hibiscus sabdariffa L.: a review, Phyther. Res. An Int. J. Devoted to Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 19 (2005) 369–375.
A.M. Osman, M. El-Garby Younes, A. Mokhtar, Sitosterol-bD-galactoside from Hibiscus sabdariffa, Phytochem. (1975).
Osman AM, Younes MeG, Mokhtar A (1975) Chemical examination of local plants. VIII. Comparative studies between constituents of different parts of Egyptian Hibiscus sabdariffa. Indian J Chem 2:1
C.T. Du, F.J. Francis, Anthocyanins of roselle (Hibiscus sabdariffa, L.), J. Food Sci. 38 (1973) 810–812.
Reaubourg G, Monceaux RH (1940) The chemical, botanical and pharmacological characteristics of the karkade (rosella) Hibiscus sabdariffa (gossypifolius). J Pharm Chim 1:292
Anon, Food Coloring Agents from Hibiscus Flowers, Patent-Japan Kokai Tokkyo Koho-81. 141 (1981) 3585.
Sato K, Goda Y, Yoshihira K, Noguchi H (1991) Structure and contents of main coloring constituents in the calyces of Hibiscus sabdariffa and commercial roselle color. Food Hyg. Saf. Sci. 32:301–307
Blunden G, Patel AV, Armstrong NJ, Gorham J (2001) Betaine distribution in the Malvaceae. Phytochemistry 58:451–454
Al-Wandawi H, Al-Shaikhly K, Abdul-Rahman M (1984) Roselle seeds: a new protein source. J Agric Food Chem 32:510–512
D.W. Bishay, C.S. Gomaa, Comparative chromatographic studies of oils of some medicinal seeds [Cassia acutifolia, C. obovata, Fenugreek, Hibiscus sabdariffa, Colchicum automnale, Citrullus colocynth; Egypt], Egypt. J. Pharm. Sci. (1976).
M.S. Karawya, M.G. Ghourab, I.M. El Shami, Study of anthocyanin content of karkadeh, Hibiscus sabdariffa [Egypt]., Egypt. J. Pharm. Sci. (1975).
Ahmad MU, Husain SK, Ahmad I, Osman SM (1979) Hibiscus sabdariffa seed oil: a re-investigation. J Sci Food Agric 30:424–428
Hassan STS, Švajdlenka E (2017) Biological evaluation and molecular docking of protocatechuic acid from Hibiscus sabdariffa L. as a potent urease inhibitor by an ESI-MS based method. Molecules 22:1696–1699
Rosa RM, Moura DJ, Melecchi MIS, dos Santos RS, Richter MF, Camarao EB, Henriques JAP, de Paula Ramos ALL, Saffi J (2007) Protective effects of Hibiscus tiliaceus L. methanolic extract to V79 cells against cytotoxicity and genotoxicity induced by hydrogen peroxide and tert-butyl-hydroperoxide. Toxicol. Vitr. 21:1442–1452
Ali M, Alam P, Singh V, Jameel M, Sultana S (2017) Phytochemical investigations of the leaves and flowers of Hibiscus rosa-sinensis L. Indian Drugs 54:30–37
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
März AM, Fabian A-K, Kozany C, Bracher A, Hausch F (2013) Large FK506-binding proteins shape the pharmacology of rapamycin. Mol Cell Biol 33:1357–1367. https://doi.org/10.1128/MCB.00678-12
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B (2021) PubChem in 2011:new data content and improved web interfaces. Nucleic Acids Res 49(2021):D1388–D1395. https://doi.org/10.1093/nar/gkaa971
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082. https://doi.org/10.1093/nar/gkx1037
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminform 3:1–14. https://doi.org/10.1186/1758-2946-3-33
Chemaxon, Chemicalize was used for [prediction of microspecies distribution] (2022). https://chemicalize.com/app/calculation.
Harding AP, Wedge DC, Popelier PLA (2009) p K a prediction from “quantum chemical topology” descriptors. J Chem Inf Model 49:1914–1924. https://doi.org/10.1021/ci900172h
Settimo L, Bellman K, Knegtel R (2014) Comparison of the accuracy of experimental and predicted pKa values of basic and acidic compounds. Pharm Res 31:1082–1095. https://doi.org/10.1007/s11095-013-1232-z
Rasul HO, Aziz BK, Ghafour DD, Kivrak A (2022) In silico molecular docking and dynamic simulation of eugenol compounds against breast cancer. J Mol Model 28:1–18. https://doi.org/10.1007/s00894-021-05010-w
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40:82–92
Molinspiration Cheminformatics, Nov. Ulica. (n.d.) Slovak Republic. https://www.molinspiration.com/.
Dong J, Wang N-N, Yao Z-J, Zhang L, Cheng Y, Ouyang D, Lu A-P, Cao D-S (2018) ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminform 10:1–11. https://doi.org/10.1186/s13321-018-0283-x
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab255
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Lipinski CA (2000) Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44:235–249. https://doi.org/10.1016/S1056-8719(00)00107-6
Zhu H, Martin TM, Ye L, Sedykh A, Young DM, Tropsha A (2009) Quantitative structure−activity relationship modeling of rat acute toxicity by oral exposure. Chem Res Toxicol 22:1913–1921. https://doi.org/10.1021/tx900189p
SeeSAR version 12.0.1, BioSolveIT GmbH, Sankt Augustin, Germany, 2022. www.biosolveit.de/SeeSAR (2022). www.biosolveit.de/SeeSAR.
Schneider N, Hindle S, Lange G, Klein R, Albrecht J, Briem H, Beyer K, Claußen H, Gastreich M, Lemmen C (2012) Substantial improvements in large-scale redocking and screening using the novel HYDE scoring function. J Comput Aided Mol Des 26:701–723
Schneider N, Lange G, Hindle S, Klein R, Rarey M (2013) A consistent description of HYdrogen bond and DEhydration energies in protein–ligand complexes: methods behind the HYDE scoring function. J Comput Aided Mol Des 27:15–29
Schärfer C, Schulz-Gasch T, Ehrlich H-C, Guba W, Rarey M, Stahl M (2013) Torsion angle preferences in druglike chemical space: a comprehensive guide. J Med Chem 56:2016–2028
S. Release, 3: Desmond molecular dynamics system, DE Shaw research, New York, NY, 2017, Maest. Interoperability Tools. Schrödinger, New York (2017).
Rasul HO, Aziz BK, Ghafour DD, Kivrak A (2022) Discovery of potential mTOR inhibitors from Cichorium intybus to find new candidate drugs targeting the pathological protein related to the breast cancer: an integrated computational approach. Mol Divers. https://doi.org/10.1007/s11030-022-10475-9
Ding Y, Fang Y, Moreno J, Ramanujam J, Jarrell M, Brylinski M (2016) Assessing the similarity of ligand binding conformations with the Contact Mode Score. Comput Biol Chem 64:403–413
García-Godoy MJ, López-Camacho E, García-Nieto J, Nebro AJ, Aldana-Montes JF (2016) Molecular docking optimization in the context of multi-drug resistant and sensitive EGFR mutants. Molecules 21:1575
Hodgson J (2001) ADMET—turning chemicals into drugs. Nat Biotechnol 19:722–726. https://doi.org/10.1038/90761
Beig A, Agbaria R, Dahan A (2013) Oral delivery of lipophilic drugs: the tradeoff between solubility increase and permeability decrease when using cyclodextrin-based formulations. PLoS ONE 8:e68237. https://doi.org/10.1371/journal.pone.0068237
Tsaioun K, Kates SA (2011) ADMET for medicinal chemists: a practical guide. Wiley, New York
Wu Z, Lei T, Shen C, Wang Z, Cao D, Hou T (2019) ADMET evaluation in drug discovery. 19. Reliable prediction of human cytochrome P450 inhibition using artificial intelligence approaches. J Chem Inf Model 59:4587–4601. https://doi.org/10.1021/acs.jcim.9b00801
Reulecke I, Lange G, Albrecht J, Klein R, Rarey M (2008) Towards an integrated description of hydrogen bonding and dehydration: decreasing false positives in virtual screening with the HYDE scoring function. ChemMedChem Chem Enabling Drug Discov 3:885–897
Martínez L (2015) Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 10:e0119264. https://doi.org/10.1371/journal.pone.0119264
Alnajjar R, Mostafa A, Kandeil A, Al-Karmalawy AA (2020) Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon 6:e05641. https://doi.org/10.1016/j.heliyon.2020.e05641
Acknowledgements
The researchers appreciate the support and resources provided by Charmo University and Komar University of Science and Technology in completing this study.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
HOR: Conceptualization, Methodology, Software, Data Curation, and Writing. BKA: Supervision, Software, and Validation. DDG: Writing and Editing, Visualization, and Resources. AK: Validation, Writing, and Editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest declared by the authors.
Ethical approval
This article does not contain any studied with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rasul, H.O., Aziz, B.K., Ghafour, D.D. et al. Screening the possible anti-cancer constituents of Hibiscus rosa-sinensis flower to address mammalian target of rapamycin: an in silico molecular docking, HYDE scoring, dynamic studies, and pharmacokinetic prediction. Mol Divers 27, 2273–2296 (2023). https://doi.org/10.1007/s11030-022-10556-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10556-9