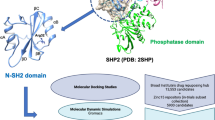
Abstract
FBXW8 plays an irreplaceable role in the substrate recognition of ubiquitin-dependent proteolysis, which further regulates cell cycle progression and signal transduction. However, the abnormal expression of FBXW8 triggers malignancy, inflammation, and autophagy irregulation. FBXW8 is considered as an effective therapeutic target for Cullin-RING ligase 7 (CRL7)-related cancers. Still, the lack of selective inhibitors hinders further therapeutic development and limits the exploration of its biological mechanism. This study constructed an integrated protocol that combines pharmacophore modeling, structure-based virtual screening, and Molecular Dynamic Simulation. It was then used as a screening query to identify hit compounds targeted at the substrate recognition site of FBXW8 from a large-scale compound database including 120 million compounds. Then, ten lead compounds were retrieved by using molecular docking analysis and ADMET prediction. Finally, MD simulations were performed to validate the binding stability of selected drug candidates. The result indicated that three newly obtained compounds, namely ZINC96179876, ZINC72174069, and ZINC97730272, might be potent FBXW8 inhibitors against CRL7-related cancers such as endometrial cancer.
Graphical abstract







Similar content being viewed by others
Abbreviations
- CRL7:
-
Cullin-RING ligase 7
- FBXW8:
-
F-box/WD repeat-containing protein 8
- CCND1:
-
Cyclin D1
- OCT4:
-
The octamer binding transcription factor 4
- MD:
-
Molecular dynamics
- HPK1:
-
Mitogen-activated protein kinase 1
References
Moutinho C, Esteller M (2017) MicroRNAs and epigenetics. Adv Cancer Res 135:189–220. https://doi.org/10.1016/bs.acr.2017.06.003
Liu X, Zhao B, Sun L, Bhuripanyo K, Wang Y, Bi Y, Davuluri RV, Duong DM, Nanavati D, Yin J, Kiyokawa H (2017) Orthogonal ubiquitin transfer identifies ubiquitination substrates under differential control by the two ubiquitin activating enzymes. Nat Commun 8:14286. https://doi.org/10.1038/ncomms14286
Li Y, Xie P, Lu L, Wang J, Diao L, Liu Z, Guo F, He Y, Liu Y, Huang Q, Liang H, Li D, He F (2017) An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat Commun 8(1):347. https://doi.org/10.1038/s41467-017-00299-9
Zheng N, Shabek N (2017) Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem 86:129–157. https://doi.org/10.1146/annurev-biochem-060815-014922
Shi W, Feng L, Dong S, Ning Z, Hua Y, Liu L, Chen Z, Meng Z (2020) FBXL6 governs c-MYC to promote hepatocellular carcinoma through ubiquitination and stabilization of HSP90AA1. Cell Commun Signal 18(1):100. https://doi.org/10.1186/s12964-020-00604-y
Nakayama KI, Nakayama K (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 6(5):369–381. https://doi.org/10.1038/nrc1881
Wang Z, Liu P, Inuzuka H, Wei W (2014) Roles of F-box proteins in cancer. Nat Rev Cancer 14(4):233–247. https://doi.org/10.1038/nrc3700
Xu C, Min J (2011) Structure and function of WD40 domain proteins. Protein Cell 2(3):202–214. https://doi.org/10.1007/s13238-011-1018-1
Islam S, Dutta P, Chopra K, Sahay O, Rapole S, Chauhan R, Santra MK (2021) Co-operative binding of SKP1, Cullin1 and Cullin7 to FBXW8 results in Cullin1-SKP1-FBXW8-Cullin7 functional complex formation that monitors cellular function of beta-TrCP1. Int J Biol Macromol 190:233–243. https://doi.org/10.1016/j.ijbiomac.2021.08.195
Yeh C-H, Bellon M, Nicot C (2018) FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. https://doi.org/10.1186/s12943-018-0857-2
Li DZ, Liu SF, Zhu L, Wang YX, Chen YX, Liu J, Hu G, Yu X, Li J, Zhang J, Wu ZX, Lu H, Liu W, Liu B (2017) FBXW8-dependent degradation of MRFAP1 in anaphase controls mitotic cell death. Oncotarget 8(57):97178–97186
Litterman N, Ikeuchi Y, Gallardo G, O’Connell BC, Sowa ME, Gygi SP, Harper JW, Bonni A (2011) An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol 9(5):e1001060. https://doi.org/10.1371/journal.pbio.1001060
Kim SJ, DeStefano MA, Oh WJ, Wu CC, Vega-Cotto NM, Finlan M, Liu D, Su B, Jacinto E (2012) mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Mol Cell 48(6):875–887. https://doi.org/10.1016/j.molcel.2012.09.029
Yoon MS (2017) The role of mammalian target of Rapamycin (mTOR) in insulin signaling. Nutrients. https://doi.org/10.3390/nu9111176
Ponyeam W, Hagen T (2012) Characterization of the Cullin7 E3 ubiquitin ligase–heterodimerization of cullin substrate receptors as a novel mechanism to regulate cullin E3 ligase activity. Cell Signal 24(1):290–295. https://doi.org/10.1016/j.cellsig.2011.08.020
Lin P, Fu J, Zhao B, Lin F, Zou H, Liu L, Zhu C, Wang H, Yu X (2011) Fbxw8 is involved in the proliferation of human choriocarcinoma JEG-3 cells. Mol Biol Rep 38(3):1741–1747. https://doi.org/10.1007/s11033-010-0288-7
Wang Z, Inuzuka H, Fukushima H, Wan L, Gao D, Shaik S, Sarkar FH, Wei W (2011) Emerging roles of the FBW7 tumour suppressor in stem cell differentiation. EMBO Rep 13(1):36–43. https://doi.org/10.1038/embor.2011.231
Wang H, Chen Y, Lin P, Li L, Zhou G, Liu G, Logsdon C, Jin J, Abbruzzese JL, Tan TH, Wang H (2014) The CUL7/F-box and WD repeat domain containing 8 (CUL7/Fbxw8) ubiquitin ligase promotes degradation of hematopoietic progenitor kinase 1. J Biol Chem 289(7):4009–4017. https://doi.org/10.1074/jbc.M113.520106
Shi D, Zhang Y, Lu R, Zhang Y (2018) The long non-coding RNA MALAT1 interacted with miR-218 modulates choriocarcinoma growth by targeting Fbxw8. Biomed Pharmacother 97:543–550. https://doi.org/10.1016/j.biopha.2017.10.083
Lan R, Jin B, Liu YZ, Zhang K, Niu T, You Z (2020) Genome and transcriptome profiling of FBXW family in human prostate cancer. Am J Clin Exp Urol 8(4):116–128
Zheng Z, Hong D, Zhang X, Chang Y, Sun N, Lin Z, Li H, Huang S, Zhang R, Xie Q, Huang H, Jin H (2021) uc.77-downregulation promotes colorectal cancer cell proliferation by inhibiting FBXW8-mediated CDK4 protein degradation. Front Oncol 11:673223. https://doi.org/10.3389/fonc.2021.673223
Webb B, Sali A (2016) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. https://doi.org/10.1002/cpbi.3
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4):486–501. https://doi.org/10.1107/S0907444910007493
Guiley KZ, Stevenson JW, Lou K, Barkovich KJ, Kumarasamy V, Wijeratne TU, Bunch KL, Tripathi S, Knudsen ES, Witkiewicz AK, Shokat KM, Rubin SM (2019) p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science. https://doi.org/10.1126/science.aaw2106
Lee H, Heo L, Lee MS, Seok C (2015) GalaxyPepDock: a protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res 43(W1):W431-435. https://doi.org/10.1093/nar/gkv495
Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z (2014) ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30(12):1771–1773. https://doi.org/10.1093/bioinformatics/btu097
Okabe H, Lee SH, Phuchareon J, Albertson DG, McCormick F, Tetsu O (2006) A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS ONE 1:e128. https://doi.org/10.1371/journal.pone.0000128
Bae KB, Yu DH, Lee KY, Yao K, Ryu J, Lim DY, Zykova TA, Kim MO, Bode AM, Dong Z (2017) Serine 347 phosphorylation by JNKs negatively regulates OCT4 protein stability in mouse embryonic stem cells. Stem Cell Rep 9(6):2050–2064. https://doi.org/10.1016/j.stemcr.2017.10.017
Sterling T, Irwin JJ (2015) ZINC 15–ligand discovery for everyone. J Chem Inf Model 55(11):2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. https://doi.org/10.1038/srep42717
Banerjee P, Eckert AO, Schrey AK, Preissner R (2018) ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res 46(W1):W257–W263. https://doi.org/10.1093/nar/gky318
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49(W1):W5–W14. https://doi.org/10.1093/nar/gkab255
Schyman P, Liu R, Desai V, Wallqvist A (2017) vNN web server for ADMET predictions. Front Pharmacol 8:889. https://doi.org/10.3389/fphar.2017.00889
Lagorce D, Bouslama L, Becot J, Miteva MA, Bo V (2017) FAF-Drugs4 free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 33(22):3658–3660. https://doi.org/10.1093/bioinformatics/xxxxx
Pires DE, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58(9):4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. https://doi.org/10.1002/jcc.20291
Vanommeslaeghe K, MacKerell AD Jr (2012) Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J Chem Inf Model 52(12):3144–3154. https://doi.org/10.1021/ci300363c
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19(8):649–658. https://doi.org/10.1016/j.neo.2017.05.002
Shi L, Du D, Peng Y, Liu J, Long J (2020) The functional analysis of Cullin 7 E3 ubiquitin ligases in cancer. Oncogenesis 9(10):98. https://doi.org/10.1038/s41389-020-00276-w
Ramachandran S, Kota P, Ding F, Dokholyan NV (2011) Automated minimization of steric clashes in protein structures. Proteins 79(1):261–270. https://doi.org/10.1002/prot.22879
Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486
Acknowledgements
This research work was supported by the Doctoral Scientific Fund Project of Henan University of Technology (2019BS050). The Student Academic and Technological Innovation Training Program (GJXY202219) contributed to the research presented in this study. Mr. Sheng Wen and Mr. Yibo Liu are thanked for their technical assistance in preparing figures.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by YZ, LC, WC, and YS. The first draft of the manuscript was written by YZ and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Cui, L., Chen, W. et al. Structure-based drug design of potential inhibitors of FBXW8, the substrate recognition component of Cullin-RING ligase 7. Mol Divers 27, 2257–2271 (2023). https://doi.org/10.1007/s11030-022-10554-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10554-x