Abstract
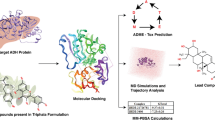
Nicotinamide N-methyltransferase (NNMT) is a protein coding gene, which methylates the nicotinamide (NA) (vitamin B3) to produce 1-methylnicotinamide (MNA). Several studies have suggested that the overexpression of NNMT is associated with different metabolic disorders like obesity and type-2 diabetes thereby making it an important therapeutic target for development of anti-diabetic agents. Here we describe a workflow for identification of new inhibitors of NNMT from a library of small molecules. In this study, we have hypothesized a four-point pharmacophore model based on the pharmacophoric features of reported NNMT inhibitors in the literature. The statistically significant pharmacophore hypothesis was used to explore the Maybridge compound library that resulted in mapping of 1330 hit compounds on the proposed hypothesis. Subsequently, a total of eight high scoring compounds, showing good protein–ligand interactions in the molecular docking study, were selected for biological evaluation of NNMT activity. Eventually, four compounds were found to show significant inhibitory activity for NNMT and can be further explored to design new derivatives around the identified scaffolds with improved activities as NNMT inhibitors.
Graphical abstract











Similar content being viewed by others
References
Aksoy S, Szumlanski CL, Weinshilboum RM (1994) Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J Biol Chem 269:14835–14840. https://doi.org/10.1016/s0021-9258(17)36700-5
Gao Y, Van Haren MJ, Moret EE et al (2019) Bisubstrate inhibitors of nicotinamide N-methyltransferase (NNMT) with enhanced activity. J Med Chem 62:6597–6614. https://doi.org/10.1021/acs.jmedchem.9b00413
Babault N, Allali-Hassani A, Li F et al (2018) Discovery of bisubstrate inhibitors of nicotinamide N-methyltransferase (NNMT). J Med Chem 61:1541–1551. https://doi.org/10.1021/acs.jmedchem.7b01422
Houtkooper RH, Cantó C, Wanders RJ, Auwerx J (2010) The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31:194–223. https://doi.org/10.1210/er.2009-0026
Ramsden DB, Waring RH, Barlow DJ, Parsons RB (2017) Nicotinamide N-methyltransferase in health and cancer. Int J Tryptophan Res. https://doi.org/10.1177/1178646917691739
Kim J, Hong SJ, Lim EK et al (2009) Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J Exp Clin Cancer Res 28:1–9. https://doi.org/10.1186/1756-9966-28-20
Riederer M, Erwa W, Zimmermann R et al (2009) Adipose tissue as a source of nicotinamide N-methyltransferase and homocysteine. Atherosclerosis 204:412–417. https://doi.org/10.1016/j.atherosclerosis.2008.09.015
Seifert R (1984) Nicotinamide methylation tissue distribution, developmental and neoplastic changes. Biochimica et Biophysica Acta (BBA) Gen Subj 801:259–264
Ridgeline Therapeutics—new mechanism-of-action drugs—ridgeline therapeutics—new drugs for widespread diseases. http://www.ridgelinetherapeutics.com/our-science.html. Accessed 26 May 2022
Ulanovskaya OA, Zuhl AM, Cravatt BF (2013) NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol 9:300–306. https://doi.org/10.1038/nchembio.1204
Hong S, Moreno-Navarrete JM, Wei X et al (2015) Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nat Med 21:887–894. https://doi.org/10.1038/nm.3882
Sperber H, Mathieu J, Wang Y et al (2015) The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol 17:1523–1535. https://doi.org/10.1038/ncb3264
Jung J, Kim LJ, Wang X et al (2017) Nicotinamide metabolism regulates glioblastoma stem cell maintenance. JCI Insight. https://doi.org/10.1172/jci.insight.90019
Policarpo RL, Decultot L, May E et al (2019) High-affinity alkynyl bisubstrate inhibitors of nicotinamide N-methyltransferase (NNMT). ChemRxiv
Van Haren MJ, Sastre Toraño J, Sartini D et al (2016) A rapid and efficient assay for the characterization of substrates and inhibitors of nicotinamide N-methyltransferase. Biochemistry 55:5307–5315. https://doi.org/10.1021/acs.biochem.6b00733
Kannt A, Rajagopal S, Kadnur SV et al (2018) A small molecule inhibitor of nicotinamide N-methyltransferase for the treatment of metabolic disorders. Sci Rep 8:1–15. https://doi.org/10.1038/s41598-018-22081-7
Ruf S, Hallur MS, Anchan NK et al (2018) Novel nicotinamide analog as inhibitor of nicotinamide N-methyltransferase. Bioorganic Med Chem Lett 28:922–925. https://doi.org/10.1016/j.bmcl.2018.01.058
Peng Y, Sartini D, Pozzi V et al (2011) Structural basis of substrate recognition in human nicotinamide N-methyltransferase. Biochemistry 50:7800–7808. https://doi.org/10.1021/bi2007614
Gao Y, Martin NI, van Haren MJ (2021) Nicotinamide N-methyl transferase (NNMT): an emerging therapeutic target. Drug Discov Today 26:2699–2706. https://doi.org/10.1016/j.drudis.2021.05.011
Khurshid Ahmad MH (2014) Drug discovery and in silico techniques: a mini-review. Enzym Eng. https://doi.org/10.4172/2329-6674.1000123
Hollingsworth SA, Dror RO (2018) Molecular dynamics simulation for all. Neuron 99:1129–1143. https://doi.org/10.1016/j.neuron.2018.08.011
Maestro | Schrödinger. https://www.schrodinger.com/products/maestro. Accessed 24 Jan 2022
LigPrep | Schrödinger. https://www.schrodinger.com/products/ligprep. Accessed 24 Jan 2022
Banks JL, Beard HS, Cao Y et al (2005) Integrated modeling program, applied chemical theory (IMPACT). J Comput Chem 26:1752–1780. https://doi.org/10.1002/jcc.20292
Dixon SL, Smondyrev AM, Rao SN (2006) PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des 67:370–372. https://doi.org/10.1111/j.1747-0285.2006.00384.x
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55:6582–6594. https://doi.org/10.1021/jm300687e
Bhagwati S, Siddiqi MI (2020) Identification of potential soluble epoxide hydrolase (sEH) inhibitors by ligand-based pharmacophore model and biological evaluation. J Biomol Struct Dyn 38:4956–4966. https://doi.org/10.1080/07391102.2019.1691659
Friesner RA, Murphy RB, Repasky MP et al (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177–6196. https://doi.org/10.1021/jm051256o
Neelakantan H, Wang HY, Vance V et al (2017) Structure-activity relationship for small molecule inhibitors of nicotinamide N-methyltransferase. J Med Chem 60:5015–5028. https://doi.org/10.1021/acs.jmedchem.7b00389
Van Haren MJ, Taig R, Kuppens J et al (2017) Inhibitors of nicotinamide: N -methyltransferase designed to mimic the methylation reaction transition state. Org Biomol Chem 15:6656–6667. https://doi.org/10.1039/c7ob01357d
Abraham MJ, Murtola T, Schulz R et al (2015) Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
SwissParam—Topology and parameters for small organic molecules. https://www.swissparam.ch/. Accessed 24 Jan 2022
Hammer Ø (2009) GROMACS reference manual. Palaeontol Electron 1–168
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Quimque MTJ, Notarte KIR, Fernandez RAT et al (2021) Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J Biomol Struct Dyn 39:4316–4333. https://doi.org/10.1080/07391102.2020.1776639
Acknowledgements
UK and LP acknowledge University Grant Commission (UGC), New Delhi, India, for providing fellowship, Chemical repository of CSIR-Central Drug Research Institute is acknowledged for providing Compounds from Maybridge library for in vitro biological evaluation. Department of Biotechnology (DBT), Ministry of Science and Technology Government of India (Grant No. GAP0384) is also gratefully acknowledged for funding this work. This manuscript has CSIR-CDRI manuscript number 10425.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kushavah, U., Panigrahi, L., Ahmed, S. et al. Ligand-based in silico identification and biological evaluation of potential inhibitors of nicotinamide N-methyltransferase. Mol Divers 27, 1255–1269 (2023). https://doi.org/10.1007/s11030-022-10485-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10485-7