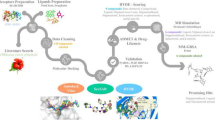
Abstract
Breast cancer is the most common malignancy among women. It is a complex condition with many subtypes based on the hormone receptor. The mammalian target of the rapamycin (mTOR) pathway regulates cell survival, metabolism, growth, and protein synthesis in response to upstream signals in both normal physiological and pathological situations, primarily in cancer. The objective of this study was to screen for a potential target to inhibit the mTOR using a variety of inhibitors derived from Cichorium intybus and to identify the one with the highest binding affinity for the receptor protein. Initially, AutoDock Vina was used to perform structure-based virtual screening, as protein-like interactions are critical in drug development. For the comparative analysis, 110 components of Cichorium intybus were employed and ten FDA-approved anticancer medicines, including everolimus, an mTOR inhibitor. Further, the drug-likeness and ADMET properties were investigated to evaluate the anti-breast cancer activity by applying Lipinski's rule of five to the selected molecules. The promising candidates were then subjected to three replica molecular dynamics simulations run for 100 ns, followed by binding free energy estimation using MM-GBSA. The data were analyzed using root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and protein–ligand interactions to determine the stability of the protein–ligand complex. Based on the results, taraxerone (98) revealed optimum binding affinities with mTOR, followed by stigmasterol (110) and rutin (104), which compared favorably to the control compounds. Subsequently, bioactive compounds derived from Cichorium intybus may serve as lead molecules for developing potent and effective mTOR inhibitors to treat breast cancer.
Graphical abstract















Similar content being viewed by others
Availability of data and material
All data generated during this study are included in supplementary information files.
Code availability
Not applicable.
References
Siegel R, DeSantis C, Jemal A (2014) Colorectal cancer statistics, 2014. CA Cancer J Clin 64:104–117. https://doi.org/10.3322/caac.21220
Chan S, Scheulen ME, Johnston S et al (2005) Phase II study of temsirolimus (CCI-779), a novel inhibitor of mTOR, in heavily pretreated patients with locally advanced or metastatic breast cancer. J Clin Oncol 23:5314–5322. https://doi.org/10.1200/JCO.2005.66.130
Castellanos M, Gubern C, Kadar E (2016) mTOR: exploring a new potential therapeutic target for stroke. In: Molecules to medicine with mTOR. Elsevier, pp 105–122
Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149:274–293. https://doi.org/10.1016/j.cell.2012.03.017
Zou Z, Tao T, Li H, Zhu X (2020) mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci 10:1–11. https://doi.org/10.1186/s13578-020-00396-1
Unni N, Arteaga CL (2019) Is dual mTORC1 and mTORC2 therapeutic blockade clinically feasible in cancer? JAMA Oncol 5:1564–1565. https://doi.org/10.1001/jamaoncol.2019.2525
Engelman JA, Luo J, Cantley LC (2006) The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7:606–619. https://doi.org/10.1038/nrg1879
Samuels Y, Wang Z, Bardelli A, et al (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science (80-). https://doi.org/10.1126/science.1096502
Gera J, Lichtenstein A (2011) The mammalian target of rapamycin pathway as a therapeutic target in multiple myeloma. Leuk Lymphoma 52:1857–1866. https://doi.org/10.3109/10428194.2011.580478
Dowling RJO, Topisirovic I, Fonseca BD, Sonenberg N (2010) Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim Biophys Acta (BBA)-Proteins Proteom 1804:433–439. https://doi.org/10.1016/j.bbapap.2009.12.001
Vittorio S, Gitto R, Adornato I et al (2021) In silico strategy for targeting the mTOR kinase at rapamycin binding site by small molecules. Molecules 26:1103. https://doi.org/10.3390/molecules26041103
Zulkipli NN, Zakaria R, Long I et al (2020) In silico analyses and cytotoxicity study of Asiaticoside and Asiatic acid from Malaysian plant as potential mTOR inhibitors. Molecules 25:3991. https://doi.org/10.3390/molecules25173991
Zoncu R, Efeyan A, Sabatini DM (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol cell Biol 12:21–35. https://doi.org/10.1038/nrm3025
Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168:960–976. https://doi.org/10.1016/j.cell.2017.02.004
Mossmann D, Park S, Hall MN (2018) mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer 18:744–757. https://doi.org/10.1038/s41568-018-0074-8
Hare SH, Harvey AJ (2017) mTOR function and therapeutic targeting in breast cancer. Am J Cancer Res 7:383
Hua H, Kong Q, Zhang H et al (2019) Targeting mTOR for cancer therapy. J Hematol Oncol 12:1–19. https://doi.org/10.1186/s13045-019-0754-1
Baselga J (2011) Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist 16:12–19. https://doi.org/10.1634/theoncologist.2011-S1-12
Di Cosimo S, Baselga J (2009) Phosphoinositide 3-kinase mutations in breast cancer: a “good” activating mutation? Clin Cancer Res 15:5017–5019. https://doi.org/10.1158/1078-0432.CCR-09-1173
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A et al (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68:6084–6091. https://doi.org/10.1158/0008-5472.CAN-07-6854
Saran U, Foti M, Dufour J-F (2015) Cellular and molecular effects of the mTOR inhibitor everolimus. Clin Sci 129:895–914. https://doi.org/10.1042/CS20150149
Kuo C-T, Chen C-L, Li C-C et al (2019) Immunofluorescence can assess the efficacy of mTOR pathway therapeutic agent Everolimus in breast cancer models. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-45319-4
Ciołczyk-Wierzbicka D, Gil D, Zarzycka M, Laidler P (2020) mTOR inhibitor everolimus reduces invasiveness of melanoma cells. Hum Cell 33:88–97. https://doi.org/10.1007/s13577-019-00270-4
MacKeigan JP, Krueger DA (2015) Differentiating the mTOR inhibitors everolimus and sirolimus in the treatment of tuberous sclerosis complex. Neuro Oncol 17:1550–1559. https://doi.org/10.1093/neuonc/nov152
Tian T, Li X, Zhang J (2019) mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int J Mol Sci 20:755. https://doi.org/10.3390/ijms20030755
Street RA, Sidana J, Prinsloo G (2013) Cichorium intybus: Traditional uses, phytochemistry, pharmacology, and toxicology. Evid-Based Complement Altern Med. https://doi.org/10.1155/2013/579319
Bais HP, Ravishankar GA (2001) Cichorium intybus L–cultivation, processing, utility, value addition and biotechnology, with an emphasis on current status and future prospects. J Sci Food Agric 81:467–484. https://doi.org/10.1002/jsfa.817
Wang Y, Lin Z, Zhang B et al (2019) Cichorium intybus L. extract suppresses experimental gout by inhibiting the NF-κB and NLRP3 signaling pathways. Int J Mol Sci 20:4921. https://doi.org/10.1002/jsfa.817
Chandra S, Kumar M, Dwivedi P, Arti K (2016) Studies on industrial importance and medicinal value of chicory plant (Cichorium intybus L.). Int J Adv Res 4:1060–1071
Liu Q, Chen Y, Shen C et al (2017) Chicoric acid supplementation prevents systemic inflammation-induced memory impairment and amyloidogenesis via inhibition of NF-κB. FASEB J 31:1494–1507. https://doi.org/10.1096/fj.201601071R
El-Sayed YS, Lebda MA, Hassinin M, Neoman SA (2015) Chicory (Cichorium intybus L.) root extract regulates the oxidative status and antioxidant gene transcripts in CCl4-induced hepatotoxicity. PLoS ONE 10:e0121549. https://doi.org/10.1371/journal.pone.0121549
Süntar I, Akkol EK, Keles H et al (2012) Comparative evaluation of traditional prescriptions from Cichorium intybus L. for wound healing: stepwise isolation of an active component by in vivo bioassay and its mode of activity. J Ethnopharmacol 143:299–309. https://doi.org/10.1016/j.jep.2012.06.036
Behboodi S, Baghbani-Arani F, Abdalan S, Shandiz SAS (2019) Green engineered biomolecule-capped silver nanoparticles fabricated from Cichorium intybus extract: in vitro assessment on apoptosis properties toward human breast cancer (MCF-7) cells. Biol Trace Elem Res 187:392–402. https://doi.org/10.1007/s12011-018-1392-0
Hazra B, Sarkar R, Bhattacharyya S, Roy P (2002) Tumour inhibitory activity of chicory root extract against Ehrlich ascites carcinoma in mice. Fitoterapia 73:730–733. https://doi.org/10.1016/S0367-326X(02)00232-0
Khan MF, Nasr FA, Noman OM et al (2020) Cichorins D-F: three new compounds from Cichorium intybus and their biological effects. Molecules 25:4160. https://doi.org/10.3390/molecules25184160
Imam KMSU, Xie Y, Liu Y et al (2019) Cytotoxicity of Cichorium intybus L. metabolites. Oncol Rep 42:2196–2212. https://doi.org/10.3892/or.2019.7336
Conforti F, Ioele G, Statti GA et al (2008) Anti-proliferative activity against human tumor cell lines and toxicity test on Mediterranean dietary plants. Food Chem Toxicol 46:3325–3332. https://doi.org/10.1016/j.fct.2008.08.004
Lee K-T, Kim J-I, Park H-J et al (2000) Differentiation-inducing effect of magnolialide, a 1β-hydroxyeudesmanolide isolated from Cichorium intybus, on human leukemia cells. Biol Pharm Bull 23:1005–1007. https://doi.org/10.1248/bpb.23.1005
Abu-Dahab R, Afifi F (2007) Anti-proliferative activity of selected medicinal plants of Jordan against a breast adenocarcinoma cell line (MCF7). Sci Pharm 75:121–146. https://doi.org/10.3797/scipharm.2007.75.121
Saleem M, Abbas K, Naseer F et al (2014) Anticancer activity of n-hexane extract of Cichorium intybus on lymphoblastic leukemia cells (Jurkat cells). African J plant Sci 8:315–319. https://doi.org/10.5897/AJPS2013.1021
Nawab A, Yunus M, Mahdi AA, Gupta S (2011) Evaluation of anticancer properties of medicinal plants from the Indian sub-continent. Mol Cell Pharmacol 3:21–29
Rahimipour A, Dehghan Nayeri N, Mehrandish R, AwsatMellati A (2017) Anti-cancer activity of methanol extracts of cichoriumintybus on human breast cancer SKBR3 Cell Line. Razavi Int J Med. https://doi.org/10.5812/RIJM.38369
Esmaeilbeig M, Kouhpayeh SA, Amirghofran Z (2015) An investigation of the growth inhibitory capacity of several medicinal plants from Iran on tumor cell lines. Iran J cancer Prev. https://doi.org/10.17795/ijcp-4032
Seto M, Miyase T, Umehara K et al (1988) Sesquiterpene lactones from Cichorium endivia L. and C. intybus L. and cytotoxic activity. Chem Pharm Bull 36:2423–2429. https://doi.org/10.1248/cpb.36.2423
Carazzone C, Mascherpa D, Gazzani G, Papetti A (2013) Identification of phenolic constituents in red chicory salads (Cichorium intybus) by high-performance liquid chromatography with diode array detection and electrospray ionisation tandem mass spectrometry. Food Chem 138:1062–1071. https://doi.org/10.1016/j.foodchem.2012.11.060
Malarz J, Stojakowska A, Szneler E, Kisiel W (2013) A new neolignan glucoside from hairy roots of Cichorium intybus. Phytochem Lett 6:59–61. https://doi.org/10.1016/j.phytol.2012.10.011
Tardugno R, Pozzebon M, Beggio M et al (2018) Polyphenolic profile of Cichorium intybus L. endemic varieties from the Veneto region of Italy. Food Chem 266:175–182. https://doi.org/10.1016/j.foodchem.2018.05.085
Liu H, Wang Q, Liu Y et al (2013) Antimicrobial and antioxidant activities of Cichorium intybus root extract using orthogonal matrix design. J Food Sci 78:M258–M263. https://doi.org/10.1111/1750-3841.12040
Lou Z, Wang H, Zhu S et al (2011) Antibacterial activity and mechanism of action of chlorogenic acid. J Food Sci 76:M398–M403. https://doi.org/10.1111/j.1750-3841.2011.02213.x
Fan H, Chen J, Lv H et al (2017) Isolation and identification of terpenoids from chicory roots and their inhibitory activities against yeast α-glucosidase. Eur Food Res Technol 243:1009–1017. https://doi.org/10.1007/s00217-016-2810-1
Jancic D, Todorovic V, Sircelj H, et al (2017) Biologically active compounds and antioxidant capacity of Cichorium intybus L. leaves from Montenegro. Ital J Food Sci 29:627–643
Epure A, Pârvu AE, Vlase L et al (2021) phytochemical profile, antioxidant, cardioprotective and nephroprotective activity of Romanian chicory extract. Plants 10:64. https://doi.org/10.3390/plants10010064
Das S, Vasudeva N, Sharma S (2016) Cichorium intybus: a concise report on its ethnomedicinal, botanical, and phytopharmacological aspects. Drug Dev Ther. https://doi.org/10.4103/2394-6555.180157
Nwafor IC, Shale K, Achilonu MC (2017) Chemical composition and nutritive benefits of chicory (Cichorium intybus) as an ideal complementary and/or alternative livestock feed supplement. Sci World J. https://doi.org/10.1155/2017/7343928
WenYing G, Li J (2012) Chicory seeds: a potential source of nutrition for food and feed. J Anim Plant Sci 13:1736–1746
Nørbæk R, Nielsen K, Kondo T (2002) Anthocyanins from flowers of Cichorium intybus. Phytochemistry 60:357–359. https://doi.org/10.1016/S0031-9422(02)00055-9
Tousch D, Lajoix A-D, Hosy E et al (2008) Chicoric acid, a new compound able to enhance insulin release and glucose uptake. Biochem Biophys Res Commun 377:131–135. https://doi.org/10.1016/j.bbrc.2008.09.088
Jangra SS, Madan VK (2018) Proximate, mineral and chemical composition of different parts of chicory (Cichorium intybus L.). J Pharmacogn Phytochem 7:3311–3315
Leclercq E (1984) Determination of lactucin in roots of chicory (Cichorium intybus L.) by high-performance liquid chromatography. J Chromatogr A 283:441–444. https://doi.org/10.1016/S0021-9673(00)96288-5
Pyrek JS (1985) Sesquiterpene lactones of Cichorium intybus and Leontodon autumnalis. Phytochemistry 24:186–188. https://doi.org/10.1016/S0031-9422(00)80838-9
Cavin C, Delannoy M, Malnoe A et al (2005) Inhibition of the expression and activity of cyclooxygenase-2 by chicory extract. Biochem Biophys Res Commun 327:742–749. https://doi.org/10.1016/j.bbrc.2004.12.061
Matos MS, Anastácio JD, Allwood JW et al (2020) Assessing the intestinal permeability and anti-inflammatory potential of sesquiterpene lactones from chicory. Nutrients 12:3547. https://doi.org/10.3390/nu12113547
Kisiel W, Zielińska K (2001) Guaianolides from Cichorium intybus and structure revision of Cichorium sesquiterpene lactones. Phytochemistry 57:523–527. https://doi.org/10.1016/S0031-9422(01)00072-3
Ren Y, Zhou Y, Chen X, Ye Y (2005) Discovery, structural determination and anticancer activities of lactucinlike guaianolides. Lett Drug Des Discov 2:444–450. https://doi.org/10.2174/1570180054771581
Rees SB, Harborne JB (1985) The role of sesquiterpene lactones and phenolics in the chemical defence of the chicory plant. Phytochemistry 24:2225–2231. https://doi.org/10.1016/S0031-9422(00)83015-0
Bischoff TA, Kelley CJ, Karchesy Y et al (2004) Antimalarial activity of lactucin and lactucopicrin: sesquiterpene lactones isolated from Cichorium intybus L. J Ethnopharmacol 95:455–457. https://doi.org/10.1016/j.jep.2004.06.031Get
Malik B, Pirzadah TB, Tahir I et al (2016) Phytochemical studies on Cichorium intybus L. (CHICORY) from Kashmir Himalaya using GC-MS. J Pharm Res 10:715–726
Ravindran R, Chithra ND, Deepa PE et al (2017) In vitro effects of caffeic acid, nortriptyline, precocene I and quercetin against Rhipicephalus annulatus (Acari: Ixodidae). Exp Appl Acarol 71:183–193. https://doi.org/10.1007/s10493-017-0105-2
Li G-Y, Zheng Y-X, Sun F-Z et al (2015) In silico analysis and experimental validation of active compounds from Cichorium intybus L. ameliorating liver injury. Int J Mol Sci 16:22190–22204. https://doi.org/10.3390/ijms160922190
Prasad NR, Karthikeyan A, Karthikeyan S, Reddy BV (2011) Inhibitory effect of caffeic acid on cancer cell proliferation by oxidative mechanism in human HT-1080 fibrosarcoma cell line. Mol Cell Biochem 349:11–19. https://doi.org/10.1007/s11010-010-0655-7
Pragasam SJ, Venkatesan V, Rasool M (2013) Immunomodulatory and anti-inflammatory effect of p-coumaric acid, a common dietary polyphenol on experimental inflammation in rats. Inflammation 36:169–176. https://doi.org/10.1007/s10753-012-9532-8
Krebsky EO, Geuns JMC, De Proft M (1999) Polyamines and sterols in Cichorium heads. Phytochemistry 50:549–553. https://doi.org/10.1016/S0031-9422(98)00555-X
Bridle P, Loeffler RST, Timberlake CF, Self R (1984) Cyanidin 3-malonylglucoside in Cichorium intybus. Phytochemistry 23:2968–2969. https://doi.org/10.1016/0031-9422(84)83058-7
Mulabagal V, Wang H, Ngouajio M, Nair MG (2009) Characterization and quantification of health beneficial anthocyanins in leaf chicory (Cichorium intybus) varieties. Eur Food Res Technol 230:47–53. https://doi.org/10.1007/s00217-009-1144-7
Chen AY, Chen YC (2013) A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem 138:2099–2107. https://doi.org/10.1016/j.foodchem.2012.11.139
Zhang S, Zhao M, Bai L et al (2006) Bioactive guaianolides from siyekucai (Ixeris chinensis). J Nat Prod 69:1425–1428. https://doi.org/10.1021/np068015j
Papetti A, Mascherpa D, Carazzone C et al (2013) Identification of organic acids in Cichorium intybus inhibiting virulence-related properties of oral pathogenic bacteria. Food Chem 138:1706–1712. https://doi.org/10.1016/j.foodchem.2012.10.148
Satmbekova D, Srivedavyasasri R, Orazbekov Y et al (2018) Chemical and biological studies on Cichorium intybus L. Nat Prod Res 32:1343–1347. https://doi.org/10.1080/14786419.2017.1343319
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucl Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
März AM, Fabian A-K, Kozany C et al (2013) Large FK506-binding proteins shape the pharmacology of rapamycin. Mol Cell Biol 33:1357–1367. https://doi.org/10.1128/MCB.00678-12
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Kim S, Chen J, Cheng T et al (2021) PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 49:D1388–D1395. https://doi.org/10.1093/nar/gkaa971
Wishart DS, Feunang YD, Guo AC et al (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082. https://doi.org/10.1093/nar/gkx1037
O’Boyle NM, Banck M, James CA et al (2011) Open Babel: an open chemical toolbox. J Cheminform 3:1–14. https://doi.org/10.1186/1758-2946-3-33
Chemaxon (2022) Chemicalize was used for [prediction of microspecies distribution]. https://chemicalize.com/app/calculation
Harding AP, Wedge DC, Popelier PLA (2009) p K a prediction from “quantum chemical topology” descriptors. J Chem Inf Model 49:1914–1924. https://doi.org/10.1021/ci900172h
Settimo L, Bellman K, Knegtel R (2014) Comparison of the accuracy of experimental and predicted pKa values of basic and acidic compounds. Pharm Res 31:1082–1095. https://doi.org/10.1007/s11095-013-1232-z
Rasul HO, Aziz BK, Ghafour DD, Kivrak A (2022) In silico molecular docking and dynamic simulation of eugenol compounds against breast cancer. J Mol Model 28:1–18. https://doi.org/10.1007/s00894-021-05010-w
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40:82–92
Dong J, Wang N-N, Yao Z-J et al (2018) ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminform 10:1–11. https://doi.org/10.1186/s13321-018-0283-x
Xiong G, Wu Z, Yi J, et al (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucl Acids Res. https://doi.org/10.1093/nar/gkab255
Molinspiration Cheminformatics. In: Nov. ulica. https://www.molinspiration.com/
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Lipinski CA (2000) Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44:235–249. https://doi.org/10.1016/S1056-8719(00)00107-6
Zhu H, Martin TM, Ye L et al (2009) Quantitative structure—activity relationship modeling of rat acute toxicity by oral exposure. Chem Res Toxicol 22:1913–1921. https://doi.org/10.1021/tx900189p
Release S (2017) 3: Desmond molecular dynamics system, DE Shaw research, New York, NY, 2017. Maest Interoperability Tools, Schrödinger, New York
Hodgson J (2001) ADMET—turning chemicals into drugs. Nat Biotechnol 19:722–726. https://doi.org/10.1038/90761
Beig A, Agbaria R, Dahan A (2013) Oral delivery of lipophilic drugs: the tradeoff between solubility increase and permeability decrease when using cyclodextrin-based formulations. PLoS ONE 8:e68237. https://doi.org/10.1371/journal.pone.0068237
Tsaioun K, Kates SA (2011) ADMET for medicinal chemists: a practical guide. Wiley
Wu Z, Lei T, Shen C et al (2019) ADMET evaluation in drug discovery. 19. Reliable prediction of human cytochrome P450 inhibition using artificial intelligence approaches. J Chem Inf Model 59:4587–4601. https://doi.org/10.1021/acs.jcim.9b00801
Martínez L (2015) Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 10:e0119264. https://doi.org/10.1371/journal.pone.0119264
Alnajjar R, Mostafa A, Kandeil A, Al-Karmalawy AA (2020) Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon 6:e05641. https://doi.org/10.1016/j.heliyon.2020.e05641
Acknowledgements
The authors would like to express their gratitude to Charmo University and Komar University of Science and Technology for their ongoing assistance and facilities in carrying out this research.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Hezha O. Rasul contributed to conceptualization, methodology, software, data curation, and writing. Bakhtyar K. Aziz contributed to supervision, software, and validation Dlzar D. Ghafour contributed to writing, review and editing, visualization, resources. Arif Kivrak contributed to validation, writing–review and editing.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest declared by the authors.
Ethical approval
This article does not contain any studies with human participants or animals performed by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rasul, H.O., Aziz, B.K., Ghafour, D.D. et al. Discovery of potential mTOR inhibitors from Cichorium intybus to find new candidate drugs targeting the pathological protein related to the breast cancer: an integrated computational approach. Mol Divers 27, 1141–1162 (2023). https://doi.org/10.1007/s11030-022-10475-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10475-9