Abstract
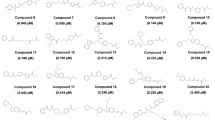
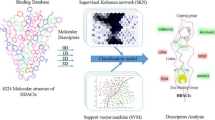
Histone deacetylase (HDAC) 1, a member of the histone deacetylases family, plays a pivotal role in various tumors. In this study, we collected 7313 human HDAC1 inhibitors with bioactivities to form a dataset. Then, the dataset was divided into a training set and a test set using two splitting methods: (1) Kohonen’s self-organizing map and (2) random splitting. The molecular structures were represented by MACCS fingerprints, RDKit fingerprints, topological torsions fingerprints and ECFP4 fingerprints. A total of 80 classification models were built by using five machine learning methods, including decision tree (DT), random forest, support vector machine, eXtreme Gradient Boosting and deep neural network. Model 15A_2 built by the XGBoost algorithm based on ECFP4 fingerprints showed the best performance, with an accuracy of 88.08% and an MCC value of 0.76 on the test set. Finally, we clustered the 7313 HDAC1 inhibitors into 31 subsets, and the substructural features in each subset were investigated. Moreover, using DT algorithm we analyzed the structure–activity relationship of HDAC1 inhibitors. It may conclude that some substructures have a significant effect on high activity, such as N-(2-amino-phenyl)-benzamide, benzimidazole, AR-42 analogues, hydroxamic acid with a middle chain alkyl and 4-aryl imidazole with a midchain of alkyl whose α carbon is chiral.
Graphical abstract




Similar content being viewed by others
References
Liu L, Dong L, Bourguet E et al (2021) Targeting class Iia HDACs: insights from phenotypes and inhibitors. Curr Med Chem 28(42):8628–8672. https://doi.org/10.2174/0929867328666210629160647
Pojani E, Barlocco D (2021) Romidepsin (FK228), A histone deacetylase inhibitor and its analogues in cancer chemotherapy. Curr Med Chem 28(7):1290–1303. https://doi.org/10.2174/0929867327666200203113926
Yang XJ, Seto E (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26(37):5310–5318. https://doi.org/10.1038/sj.onc.1210599
West AC, Johnstone RW (2014) New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 124(1):30–39. https://doi.org/10.1172/JCI69738
Gregoretti IV, Lee YM, Goodson HV (2004) Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 338(1):17–31. https://doi.org/10.1016/j.jmb.2004.02.006
Zhang B, Wang Y, Pang X (2012) Enhanced radiosensitivity of EC109 cells by inhibition of HDAC1 expression. Med Oncol 29(1):340–380. https://doi.org/10.1007/s12032-010-9559-3
Halkidou K, Gaughan L, Cook S et al (2004) Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 59(2):177–189. https://doi.org/10.1002/pros.20022
Choi JH, Kwon HJ, Yoon BI et al (2001) Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn J Caner Res 92(12):1300–1304
Zhang Z, Yamashita H, Toyama T et al (2005) Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat 94(1):11–16. https://doi.org/10.1007/s10549-005-6001-1
Ozawa A, Tanji N, Kikugawa T et al (2010) Inhibition of bladder tumour growth by histone deacetylase inhibitor. BJU Int 105(8):1181–1186. https://doi.org/10.1111/j.1464-410X.2009.08795.x
Duvic M, Vu J (2007) Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs 16(7):1111–1120. https://doi.org/10.1517/13543784.16.7.1111
VanderMolen KM, McCulloch W, Pearce CJ et al (2011) Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): a natural product recently approved for cutaneous T-cell lymphoma. J Antibiot (Tokyo) 64(8):525–531. https://doi.org/10.1038/ja.2011.35
Lee HZ, Kwitkowski VE, Del Valle PL et al (2015) FDA approval: belinostat for the treatment of patients with relapsed or refractory peripheral T-cell lymphoma. Clin Cancer Res 21(12):2666–2670. https://doi.org/10.1158/1078-0432.CCR-14-3119
Raedler LA (2016) Farydak (Panobinostat): First HDAC Inhibitor approved for patients with relapsed multiple myeloma. American Health Drug Benefits 9 (Special):84–87
Shi Y, Jia B, Xu W et al (2017) Chidamide in relapsed or refractory peripheral T cell lymphoma: a multicenter real-world study in China. J Hematol Oncol 10(1):69. https://doi.org/10.1186/s13045-017-0439-6
Remiszewski SW (2003) The Discovery of NVP-LAQ824: From Concept to Clinic. Curr Med Chem 10:2393–2402
Seki M, LaCanna R, Powers JC et al (2016) Class i histone deacetylase inhibition for the treatment of sustained atrial fibrillation. J Pharmacol Exp Ther 358(3):441–449. https://doi.org/10.1124/jpet.116.234591
Knipstein J, Gore L (2011) Entinostat for treatment of solid tumors and hematologic malignancies. Drug Evaluation 20(10):1455–1467. https://doi.org/10.1517/13543784.2011.613822
Boumber Y, Younes A, Garcia-Manero G (2011) Mocetinostat (MGCD0103): a review of an isotype-specific histone deacetylase inhibitor. Expert Opin Investig Drugs 20(6):823–829. https://doi.org/10.1517/13543784.2011.577737
Tng J, Lim J, Wu KC et al (2020) Achiral derivatives of hydroxamate AR-42 potently inhibit class I HDAC enzymes and cancer cell proliferation. J Med Chem 63(11):5956–5971. https://doi.org/10.1021/acs.jmedchem.0c00230
Lee H, Chang C, Su C et al (2016) 2-(Phenylsulfonyl)quinoline N -hydroxyacrylamides as potent anticancer agents inhibiting histone deacetylase. Eur J Med Chem 122:92–101. https://doi.org/10.1016/j.ejmech.2016.06.023
Salvador LA, Park H, Al-Awadhi FH et al (2014) Modulation of activity profiles for largazole-based HDAC inhibitors through alteration of prodrug properties. ACS Med Chem Lett 5(8):905–910. https://doi.org/10.1021/ml500170r
Mehndiratta S, Wang R, Huang H et al (2017) 4-Indolyl- N -hydroxyphenylacrylamides as potent HDAC class I and IIB inhibitors in vitro and in vivo. Eur J Med Chem 134:13–23. https://doi.org/10.1016/j.ejmech.2017.03.079
Xie R, Li Y, Tang P et al (2018) Design, synthesis and biological evaluation of novel 2-aminobenzamides containing dithiocarbamate moiety as histone deacetylase inhibitors and potent antitumor agents. Eur J Med Chem 143:320–333. https://doi.org/10.1016/j.ejmech.2017.08.041
Jones P, Altamura S, De Francesco R et al (2008) Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett 18(6):1814–1819. https://doi.org/10.1016/j.bmcl.2008.02.025
Dong G, Chen W, Wang X et al (2017) Small molecule inhibitors simultaneously targeting cancer metabolism and epigenetics: discovery of novel nicotinamide phosphoribosyltransferase (NAMPT) and Histone deacetylase (HDAC) dual inhibitors. J Med Chem 60(19):7965–7983. https://doi.org/10.1021/acs.jmedchem.7b00467
Estiu G, West N, Mazitschek R et al (2010) On the inhibition of histone deacetylase 8. Bioorg Med Chem 18(11):4103–4110. https://doi.org/10.1016/j.bmc.2010.03.080
Marson CM, Matthews CJ, Atkinson SJ et al (2015) Potent and selective inhibitors of histone deacetylase-3 containing chiral oxazoline capping groups and an-(2-aminophenyl)-benzamide binding unit. J Med Chem 58(17):6803–6818. https://doi.org/10.1021/acs.jmedchem.5b00545
Tian Y, Zhang S, Yin H et al (2020) Quantitative structure-activity relationship (QSAR) models and their applicability domain analysis on HIV-1 protease inhibitors by machine learning methods. Chemom Intell Lab Syst 196:103888. https://doi.org/10.1016/j.chemolab.2019.103888
Guo Y, Xiao J, Guo Z et al (2005) Exploration of a binding mode of indole amide analogues as potent histone deacetylase inhibitors and 3D-QSAR analyses. Bioorg Med Chem 13(18):5424–5434. https://doi.org/10.1016/j.bmc.2005.05.016
Tang H, Wang X, S, Huang X, P, et al (2009) Novel inhibitors of human histone deacetylase (HDAC) identified by QSAR modeling of known inhibitors, virtual screening, and experimental validation. J Chem Inf Model 49(2):461–476. https://doi.org/10.1021/ci800366f
Abdizadeh T, Ghodsi R, Hadizadeh F (2017) 3D-QSAR (CoMFA, CoMSIA) and molecular docking studies on histone deacetylase 1 selective inhibitors. Recent Pat Anti Cancer Drug Dis 12(4):365–383. https://doi.org/10.2174/1574892812666170508125927
Myles AJ, Feudale RN, Liu Y et al (2004) An introduction to decision tree modeling. J Chemom 18(6):275–285. https://doi.org/10.1002/cem.873
Schonlau M, Zou RY (2020) The random forest algorithm for statistical learning. Stata J Promot Commun Stat Stata 20(1):3–29. https://doi.org/10.1177/1536867x20909688
Harrington PdB (2015) Support vector machine classification trees. Anal Chem 87(21):11065–11071. https://doi.org/10.1021/acs.analchem.5b03113
Sheridan RP, Wang WM, Liaw A et al (2016) Extreme gradient boosting as a method for quantitative structure-activity relationships. J Chem Inf Model 56(12):2353–2360. https://doi.org/10.1021/acs.jcim.6b00591
Ma J, Sheridan RP, Liaw A et al (2015) Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model 55(2):263–274. https://doi.org/10.1021/ci500747n
ChEMBL. https://www.ebi.ac.uk/chembl/. Accessed May 2022
Reaxys. https://www.reaxys.com. Accessed May 2022
SONNIA. https://www.mn-am.com/products/sonnia. Accessed May 2022
Zhang S, Li Y, Qin Z et al (2019) SAR study on inhibitors of GIIA secreted phospholipase A2 using machine learning methods. Chem Biol Drug Des 93(5):666–684. https://doi.org/10.1111/cbdd.13470
RDKit. http://www.rdkit.org. Accessed May 2022
scikit-learn. http://scikit-learn.org/stable/. Accessed May 2022
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830
Xavier MM, Heck GS, Avila MBD et al (2016) SAnDReS a computational tool for statistical analysis of docking results and development of scoring functions. Comb Chem High Throughput Screening 19(10):801–812. https://doi.org/10.2174/1386207319666160927111347
Bitencourt-Ferreira G, AzevedoJr WFd (2019) machine learning to predict binding affinity. Methods Mol Biol 2053:251–273. https://doi.org/10.1007/978-1-4939-9752-7_16
XGBoost. https://pypi.python.org/pypi/xgboost/. Accessed May 2022
Keras. https://keras.io/. Accessed May 2022
Polishchuk P (2017) Interpretation of quantitative structure-activity relationship models: past, present, and future. J Chem Inf Model 57(11):2618–2639. https://doi.org/10.1021/acs.jcim.7b00274
Qin Z, Xi Y, Zhang S et al (2019) Classification of cyclooxygenase-2 inhibitors using support vector machine and random forest methods. J Chem Inf Model 59(5):1988–2008. https://doi.org/10.1021/acs.jcim.8b00876
Kong Y, Bender A, Yan A (2018) Identification of novel aurora kinase a (aurka) inhibitors via hierarchical ligand-based virtual screening. J Chem Inf Model 58(1):36–47. https://doi.org/10.1021/acs.jcim.7b00300
Wang X, Gotoh O (2009) Accurate molecular classification of cancer using simple rules. BMC Med Genomics 2:64. https://doi.org/10.1186/1755-8794-2-64
Rahman R, Matlock K, Ghosh S et al (2017) Heterogeneity aware random forest for drug sensitivity prediction. Sci Rep 7(1):11347. https://doi.org/10.1038/s41598-017-11665-4
Chen T, Guestrin C (2016) XGBoost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, pp 785–794. Doi: https://doi.org/10.1145/2939672.2939785
Tu G, Qin Z, Huo D et al (2020) Fingerprint-based computational models of 5-lipo-oxygenase activating protein inhibitors: Activity prediction and structure clustering. Chem Biol Drug Des 96(3):931–947. https://doi.org/10.1111/cbdd.13657
Walsh I, Fishman D, Garcia-Gasulla D et al (2021) DOME: recommendations for supervised machine learning validation in biology. Nat Methods 18(10):1122–1127. https://doi.org/10.1038/s41592-021-01205-4
Lvd M, Hinton G (2008) Viualizing data using t-SNE. J Mach Learn Res 9:2579–2605
Kanungo T, Mount DM, Netantahu NS et al (2002) An Efficient k-Means Clustering Algorithm: Analysis and Implementation. IEEE Trans Pattern Anal Mach Intell 24:881–892. https://doi.org/10.1109/TPAMI.2002.1017616
Rousseeuw P (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65. https://doi.org/10.1016/0377-0427(87)90125-7
Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50(5):742–754. https://doi.org/10.1021/ci100050t
Wang H, Qin Z, Yan A (2021) Classification models and SAR analysis on CysLT1 receptor antagonists using machine learning algorithms. Mol Divers 25(3):1597–1616. https://doi.org/10.1007/s11030-020-10165-4
Murahari S, Jalkanen AL, Kulp SK et al (2017) Sensitivity of osteosarcoma cells to HDAC inhibitor AR-42 mediated apoptosis. BMC Cancer 17(1):17–67. https://doi.org/10.1186/s12885-017-3046-6
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21675010), and “Chemical Grid Project” of Beijing University of Chemical Technology. We thank Molecular Networks GmbH, Erlangen, Germany for providing the programs CORINA Symphony.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, R., Tian, Y., Yang, Z. et al. Classification models and SAR analysis on HDAC1 inhibitors using machine learning methods. Mol Divers 27, 1037–1051 (2023). https://doi.org/10.1007/s11030-022-10466-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10466-w