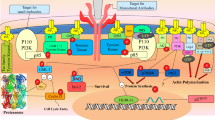
Abstract
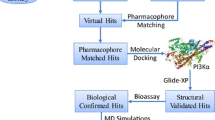
Serine/threonine-protein kinase N2 (PKN2) plays an important role in cell cycle progression, cell migration, cell adhesion and transcription activation signaling processes. In cancer, however, it plays important roles in tumor cell migration, invasion and apoptosis. PKN2 inhibitors have been shown to be promising in treating cancer. This prompted us to model this interesting target using our QSAR-guided selection of docking-based pharmacophores approach where numerous pharmacophores are extracted from docked ligand poses and allowed to compete within the context of QSAR. The optimal pharmacophore was sterically-refined, validated by receiver operating characteristic (ROC) curve analysis and used as virtual search query to screen the National Cancer Institute (NCI) database for new promising anti-PKN2 leads of novel chemotypes. Three low micromolar hits were identified with IC50 values ranging between 9.9 and 18.6 µM. Pharmacological assays showed promising cytotoxic properties for active hits in MTT and wound healing assays against MCF-7 and PANC-1 cancer cells.
Graphical abstract










Similar content being viewed by others
Abbreviations
- PKN2:
-
Protein kinase N2
- ATP:
-
Adenosine triphosphate
- QSAR:
-
Quantitative structure activity relationship
- ROC:
-
Receiver operating characteristic
- IC50 :
-
Inhibitor concentration that causes 50% enzyme inhibition
- MTT:
-
4,5-Dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide
- GA/MLR:
-
Genetic algorithm/Multiple linear regression
- AUC:
-
Area under the curve
- Hbic:
-
Hydrophobic
- HBA:
-
Hydrogen bond acceptor
- HBD:
-
Hydrogen bond donor
- PosIon:
-
Positive ionizable feature
- SNC:
-
Sensitivity
- SPC:
-
Specificity
- ACC:
-
Accuracy
- MCF7:
-
Breast cancer cell line
References
Danno S, Kubouchi K, Mehruba M, Abe M, Natsume R, Sakimura K, Eguchi S, Oka M, Hirashima M, Yasuda H, Mukai H (2017) PKN2 is essential for mouse embryonic development and proliferation of mouse fibroblasts. Gen Cells 22(2):220–236. https://doi.org/10.1111/gtc.12470
Mukai H, Muramatsu A, Mashud R, Kubouchi K, Tsujimoto S, Hongu T, Kanaho Y, Tsubaki M, Nishida S, Shioi G, Danno S, Mehruba M, Satoh R, Sugiura R (2016) PKN3 is the major regulator of angiogenesis and tumor metastasis in mice. Sci Rep 6(1):18979. https://doi.org/10.1038/srep18979
Quilliam LA, Lambert QT, Mickelson-Young LA, Westwick JK, Sparks AB, Kay BK, Jenkins NA, Gilbert DJ, Copeland NG, Der CJ (1996) Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem 271(46):28772–28776. https://doi.org/10.1074/jbc.271.46.28772
Vincent S, Settleman J (1997) The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol Cell Biol 17(4):2247–2256. https://doi.org/10.1128/mcb.17.4.2247
Hutchinson CL, Lowe PN, McLaughlin SH, Mott HR, Owen D (2013) Differential binding of RhoA, RhoB, and RhoC to protein kinase C-related kinase (PRK) isoforms PRK1, PRK2, and PRK3: PRKs have the highest affinity for RhoB. Biochem 52(45):7999–8011. https://doi.org/10.1021/bi401216w
Calautti E, Grossi M, Mammucari C, Aoyama Y, Pirro M, Ono Y, Li J, Dotto GP (2002) Fyn tyrosine kinase is a downstream mediator of Rho/PRK2 function in keratinocyte cell-cell adhesion. J Cell Biol 156(1):137–148. https://doi.org/10.1083/jcb.200105140
Bourguignon LY, Singleton PA, Diedrich F (2004) Hyaluronan-CD44 interaction with Rac1-dependent protein kinase N-gamma promotes phospholipase Cgamma1 activation, Ca(2+) signaling, and cortactin-cytoskeleton function leading to keratinocyte adhesion and differentiation. J Biol Chem 279(28):29654–29669. https://doi.org/10.1074/jbc.M403608200
Wallace SW, Magalhaes A, Hall A (2011) The Rho target PRK2 regulates apical junction formation in human bronchial epithelial cells. Mol cell biol 31(1):1–91. https://doi.org/10.1128/mcb.01001-10
Lachmann S, Jevons A, De RM, Casamassima A, Radtke S, Collazos A, Parker PJ (2011) Regulatory domain selectivity in the cell-type specific PKN-dependence of cell migration. PLoS ONE 6(7):e21732. https://doi.org/10.1371/journal.pone.0021732
Yang CS, Melhuish TA, Spencer A, Ni L, Hao Y, Jividen K, Harris TE, Snow C, Frierson HF Jr, Wotton D, Paschal BM (2017) The protein kinase C super-family member PKN is regulated by mTOR and influences differentiation during prostate cancer progression. Prostate 77(15):1452–1467
Schmidt A, Durgan J, Magalhaes A, Hall A (2007) Rho GTPases regulate PRK2/PKN2 to control entry into mitosis and exit from cytokinesis. EMBO J 26(6):1624–1636. https://doi.org/10.1038/sj.emboj.7601637
Scott F, Fala AM, Pennicott LE, Reuillon TD, Massirer KB, Elkins JM, Ward SE (2020) Development of 2-(4-pyridyl)-benzimidazoles as PKN2 chemical tools to probe cancer. Bioorg Med Chem Lett 30(8):127040. https://doi.org/10.1016/j.bmcl.2020.127040
Asquith CRM, Temme L, Laitinen T, Pickett J, Kwarcinski FE, Sinha P, Wells CI, Tizzard GJ, Zutshi R, Drewry DH (2020) Identification and Optimization of cell active 4-anilino-quin(az)oline Inhibitors for Protein Kinase Novel 3 (PKN3). bioRxiv, https://doi.org/10.1101/2020.03.02.972943
Huber K, Brault L, Fedorov O, Gasser C, Filippakopoulos P, Bullock AN, Fabbro D, Trappe J, Schwaller J, Knapp S, Bracher F (2012) 7,8-dichloro-1-oxo-β-carbolines as a versatile scaffold for the development of potent and selective kinase inhibitors with unusual binding modes. J Med Chem 55(1):403–413. https://doi.org/10.1021/jm201286z
Al-Shaer MA, Al-Aqtash RA, Taha MO (2019) Discovery of new Phosphoinositide 3-kinase delta (PI3Kdelta) inhibitors via virtual screening using crystallography-derived pharmacophore modelling and QSAR analysis. Med Chem 15(6):588–601. https://doi.org/10.2174/1573406415666190222125333
Mousa LA, Hatmal MM, Taha M (2022) Exploiting activity cliffs for building pharmacophore models and comparison with other pharmacophore generation methods: sphingosine kinase 1 as case study. J Comput Aided Mol Des 36(1):39–62. https://doi.org/10.1007/s10822-021-00435-0
Tuffaha GO, Hatmal MM, Taha MO (2019) Discovery of new JNK3 inhibitory chemotypes via QSAR-Guided selection of docking-based pharmacophores and comparison with other structure-based pharmacophore modeling methods. J Mol Graph Model 91:30–51. https://doi.org/10.1016/j.jmgm.2019.05.015
Hatmal MM, Abuyaman O, Taha M (2021) Docking-generated multiple ligand poses for bootstrapping bioactivity classifying Machine Learning: Repurposing covalent inhibitors for COVID-19-related TMPRSS2 as case study. Comput Struct Biotechnol J 19:4790–4824. https://doi.org/10.1016/j.csbj.2021.08.023
Abuhamdah S, Habash M, Taha MO (2013) Elaborate ligand-based modeling coupled with QSAR analysis and in silico screening reveal new potent acetylcholinesterase inhibitors. J Comput Aided Mol Des 27(12):1075–1092. https://doi.org/10.1007/s10822-013-9699-6
Abuhammad A, Taha M (2016) Innovative computer-aided methods for the discovery of new kinase ligands. Future Med Chem 8(5):509–526. https://doi.org/10.4155/fmc-2015-0003
Al-Nadaf AH, Taha MO (2011) Discovery of new renin inhibitory leads via sequential pharmacophore modeling, QSAR analysis, in silico screening and in vitro evaluation. J Mol Graph Model 29(6):843–864. https://doi.org/10.1016/j.jmgm.2011.02.001
Al-Sha’er MA, Taha MO (2010) Elaborate ligand-based modeling reveals new nanomolar heat shock protein 90α inhibitors. J Chem Inf Model 50(9):1706–1723. https://doi.org/10.1021/ci100222k
Al-Sha’er MA, VanPatten S, Al-Abed Y, Taha MO (2013) Elaborate ligand-based modeling reveal new migration inhibitory factor inhibitors. J Mol Graph Model 42:104–114. https://doi.org/10.1016/j.jmgm.2013.03.003
Diller DJ, Merz KMJ (2001) High throughput docking for library design and library prioritization. Proteins 43(2):113–124
Rao SN, Head MS, Kulkarni A, LaLonde JM (2007) Validation studies of the site-directed docking program LibDock. J Chem Inf Model 47(6):2159–2171
Venkatachalam CM, Jiang X, Oldfield T, Waldman M (2003) LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph 21(4):289–307
Wu G, Robertson DH, Brooks CL, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER-A CHARMm-based MD docking algorithm. J Comput Chem 24(13):1549–1562
Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP (2014) The ChEMBL bioactivity database: an update. Nucl Acid Res. https://doi.org/10.1093/nar/gkt1031
Morwick T, Büttner FH, Cywin CL, Dahmann G, Hickey E, Jakes S, Young E (2010) Hit to lead account of the discovery of bisbenzamide and related ureidobenzamide inhibitors of rho kinase. J Med Chem 53(2):759–777
Shaw D, Hollingworth G, Soldermann N, Sprague E, Schuler W (2014) Novel ROCK inhibitors for the treatment of pulmonary arterial hypertension. Bioorg Med Chem Lett 24(20):4812–4817
Biovia DS (2016) Discovery Studio Modeling Environment. Biovia 4.5 ed. San Diego: Dassault Systèmes
Lu B, Wong CF, McCammon JA (2005) Release of ADP from the catalytic subunit of protein kinase A: a molecular dynamics simulation study Release of ADP from the catalytic subunit of protein kinase A: a molecular dynamics simulation study. Prot Science 14(1):159–168. https://doi.org/10.1110/ps.04894605
Taha MO, Habash M, Khanfar MA (2014) The use of docking-based comparative intermolecular contacts analysis to identify optimal docking conditions within glucokinase and to discover of new GK activators. J Comput Aided Mol Des 28:509–547
Gao WR, Lai YL (1998) SCORE: a new empirical method for estimating the binding affinity of a protein–ligand complex. J Mol Model 4:379–394
Krammer A, Kirchhoff PD, Jiang X, Venkatachalam CM, Waldman M (2005) LigScore: a novel scoring function for predicting binding affinities. J Mol Graph Model 23:395–407
Jain AN (2006) Scoring functions for protein-ligand docking. Curr Protein Pept Sci 7(5):407–420
Velec HFG, Gohlke H, Klebe G (2005) Drug score-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J Med Chem 48:6296–6303
Rajamani R, Good AC (2007) Ranking poses in structure-based lead discovery and optimization: current trends in scoring function development. Curr Opin Drug Discov Devel 10:308–315
Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Fogel LJ, Freer ST (1995) Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: conformationally flexible docking by evolutionary programming. Chem Biol 2(5):317–324
Muegge I, Martin YC (1999) A general and fast scoring function for protein-ligand interactions: a simplified potential approach. J Med Chem 42(5):791–804
Muegge I (2002) in A knowledge-based scoring function for protein-ligand interactions: Probing the reference state BT - Virtual Screening: An Alternative or Complement to High Throughput Screening? Proceedings of the Workshop “New Approaches in Drug Design and Discovery”, (Eds.: G. Klebe), Dordrecht, Springer Netherlands, 99–114
Momany FA, Rone R (1992) Validation of the general purpose QUANTA ®3.2/CHARMm® force field. J Comput Chem 13(7):888–900
Dannenberg JJ (1997) An introduction to hydrogen bonding by george a. jeffrey (university of pittsburgh). oxford university press: New York and Oxford.120, 22 5604–5604.
Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model 45(1):160–169
Kurogi Y, Güner OF (2001) Pharmacophore modeling and three-dimensional database searching for drug design using catalyst. Curr Med Chem 8(9):1035–1055
Rogers D, Hopfinger AJ (1994) Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci 34(4):854–866
Khanfar MA, Taha MO (2013) Elaborate ligand-based modeling coupled with multiple linear regression and k nearest neighbor QSAR analyses unveiled new nanomolar mTOR inhibitors. J Chem Inf Model 53(10):2587–2612
Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP (1997) Empirical scoring functions: I The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des 11(5):425–445
Ewing TJ, Makino S, Skillman AG, Kuntz ID (2001) DOCK 4.0: Search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des 15(5):411–428
Gehlhaar DK, Bouzida D, Rejto PA (1999) In rational drug design: novel methodology and practical applications, Ser. ACS symposium 719 (Eds.: A. L. Parrill, M. R. Reddy, ACS, Washington DC, 292–311
Hahn M (1997) Three-dimensional shape-based searching of conformationally flexible compounds. J Chem Inf Comput Sci 37(1):80–86
Triballeau N, Acher F, Brabet I, Pin J, Bertrand H (2005) Virtual screening workflow development guided by the “receiver operating characteristic” curve approach application to high-throughput docking on metabotropic glutamate receptor subtype 4. J Med Chem 48(7):2534–2547
Taha MO (2012) in Virtual Screening, Vol. (Eds.: M. O. Taha), Rijeka, InTech, 3–16.
Kirchmair J, Distinto S, Schuster D, Spitzer G, Langer T, Wolber G (2008) Enhancing drug discovery through in silico screening: strategies to increase true positives retrieval rates. Curr Med Chem 15(20):2040–2053
Abutayeh RF, Taha MO (2019) Discovery of novel Flt3 inhibitory chemotypes through extensive ligand-based and new structure-based pharmacophore modelling methods. J Mol Graph Model 88:128–151
Ma H, Deacon S, Horiuchi K (2008) The challenge of selecting protein kinase assays for lead discovery optimization. Expert Opin Drug Discov 3(6):607–621
Thermofisher. Z'-LYTE® Kinase Assay . https://www.thermofisher.com/ 2021
Walters WP, Namchuk M (2003) Designing screens: how to make your hits a hit. Nat Rev Drug Discov 2(4):259–266
Zhu Q, Jiang L, Wang X (2017) The expression of Duffy antigen receptor for chemokines by epithelial ovarian cancer decreases growth potential. Oncol Lett 13:4302–4306
Pouliot N, Pearson HB (2000–2013) A. Burrows investigating metastasis using in vitro platforms. in: madame curie bioscience Database [Internet]. Austin (TX): Landes Bioscience;. Available from: https://www.ncbi.nlm.nih.gov/books/NBK100379/
Tirado-Rives J, Jorgensen WL (2006) Contribution of conformer focusing to the uncertainty in predicting free energies for protein-ligand binding. J Med Chem 49:5880–5884
Feig M, Charles L (2004) Brooks III: recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol 14:217–224
Scott F (2020) Development of PKN2 Chemical probes to enable drug discovery. Dissertation, University of Sussex
Cao DS, Xu QS, Hu QN, Liang YZ (2013) ChemoPy: freely available python package for computational biology and chemoinformatics. Bioinformatics 29(8):1092–1094
Rohrbaugh RH, Jurs PC (1987) Descriptions of molecular shape applied in studies of structure/activity and structure/property relationships. Anal Chim Acta 199:99–109
Kier LB, Hall LH (1986) Molecular Connectivity in Structure-Activity Analysis. RSP-Wiley, Chichetser (UK)
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley, Weinheim, Germany
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Protein kinase N (PKN) family: protein kinase N2. Last modified on 19/02/2015. Accessed on 29/08/2020. IUPHAR/BPS Guide to PHARMACOLOGY, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1521
Zhao J, Fang L, Zhang X, Liang Y, Gou S (2016) Synthesis and biological evaluation of new [1, 2, 4] triazolo [4, 3-a] pyridine derivatives as potential c-Met inhibitors. Bioorg med chem 24(16):3483–3493
Shoichet BK (2006) Interpreting steep dose-response curves in early inhibitor discovery. J Med Chem 49(25):7274–7277
Lachmann S, Jevons A, De Rycker M, Casamassima A, Radtke S, Collazos A, Parker PJ (2011) Regulatory domain selectivity in the cell-type specific PKN-dependence of cell migration. PLoS ONE 6:e21732
Acknowledgements
The authors thank the Deanships of Scientific Research at the Zarqa University and The University of Jordan for funding this project. We are greatly thankful for National Cancer Institute for the free NCI compounds gift. We appreciate the efforts of Ms. Haneen Sallam and Ms. Walaa Wahdan for the work in the tissue culture laboratory, help in media preparation, culturing the cancerous cells and sample preparation.
Funding
Zarqa University,6-2020,Mahmoud A Al-Sha'er
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Al-Sha’er, M.A., Basheer, H.A. & Taha, M.O. Discovery of new PKN2 inhibitory chemotypes via QSAR-guided selection of docking-based pharmacophores. Mol Divers 27, 443–462 (2023). https://doi.org/10.1007/s11030-022-10434-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10434-4