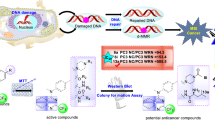
Abstract
In the past two decades, the treatment of metastatic colorectal cancer (mCRC) has been revolutionized as multiple cytotoxic, biological, and targeted drugs are being approved. Unfortunately, tumors treated with single targeted agents or therapeutics usually develop resistance. According to pathway-oriented screens, mCRC cells evade EGFR inhibition by HER2 amplification and/or activating Kras-MEK downstream signaling. Therefore, treating mCRC patients with dual EGFR/HER2 inhibitors, MEK inhibitors, or the combination of the two drugs envisaged to prevent the resistance development which eventually improves the overall survival rate. In the present study, we aimed to screen potential phytochemical lead compounds that could multi-target EGFR, HER2, and MEK1 (Mitogen-activated protein kinase kinase) using a computer-aided drug design approach that includes molecular docking, endpoint binding free energy calculation using MM-GBSA, ADMET, and molecular dynamics (MD) simulations. Docking studies revealed that, unlike all other ligands, apigenin and kaempferol exhibit the highest docking score against all three targets. Details of ADMET analysis, MM/GBSA, and MD simulations helped us to conclusively determine apigenin and kaempferol as potentially an inhibitor of EGFR, HER2, and MEK1 apigenin and kaempferol against mCRC at a systemic level. Additionally, both apigenin and kaempferol elicited antiangiogenic properties in a dose-dependent manner. Collectively, these findings provide the rationale for drug development aimed at preventing CRC rather than intercepting resistance.
Graphical Abstract








Similar content being viewed by others
Abbreviations
- CRC:
-
Colorectal cancer
- EGFR:
-
Epidermal growth factor receptor
- HER2:
-
Human epidermal growth receptor 2
- RTK:
-
Receptor tyrosine kinase
- MEK:
-
Mitogen-activated protein kinase
- MM/GBSA:
-
Molecular mechanics energies/generalized Born and surface area continuum solvation
- KMP:
-
Kaempferol
- API:
-
Apigenin
- Lap:
-
Lapatinib
- Tra:
-
Trametinib
- SMILES:
-
Simplified molecule input line entry specification
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- % HIA:
-
Human intestinal absorption (%)
- CNS permeability:
-
Central nervous system permeability
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
References
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics 2019. Cancer J Clin 69:7–34. https://doi.org/10.3322/caac.21551
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Weinstein IB (2002) Cancer: addiction to oncogenes - the achilles heal of cancer. Science 297:63–64
Misale S, di Nicolantonio F, Sartore-Bianchi A et al (2014) Resistance to Anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 4:1269–1280
Chong CR, Jänne PA (2013) The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 19:1389–1400
Dagogo-Jack I, Shaw AT (2018) Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15:81–94
Bozic I, Reiter JG, Allen B et al (2013) Evolutionary dynamics of cancer in response to targeted combination therapy. elife. https://doi.org/10.7554/eLife.00747
Stewart EL, Tan SZ, Liu G, Tsao MS (2015) Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Trans Lung Cancer Res 4:67–81
Takezawa K, Pirazzoli V, Arcila ME et al (2012) HER2 amplification: a potential mechanism of acquired resistance to egfr inhibition in EGFR -mutant lung cancers that lack the second-site EGFR T790M mutation. Cancer Discov 2:922–933. https://doi.org/10.1158/2159-8290.CD-12-0108
Rexer BN, Ghosh R, Narasanna A et al (2013) Human breast cancer cells harboring a gatekeeper T798M mutation in HER2 overexpress EGFR ligands and are sensitive to dual inhibition of EGFR and HER2. Clin Cancer Res 19:5390–5401. https://doi.org/10.1158/1078-0432.CCR-13-1038
Lenferink AEG, Pinkas-Kramarski R, van de Poll MLM et al (1998) Differential endocytic routing of homo- and hetero-dimeric ErbB tyrosine kinases confers signaling superiority to receptor heterodimers. EMBO J 17:3385–3397. https://doi.org/10.1093/emboj/17.12.3385
Graus-Porta D, Beerli RR, Daly JM, Hynes NE (1997) ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J 16:1647–1655. https://doi.org/10.1093/emboj/16.7.1647
Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2:127–137
Rowinsky EK (2004) The erbB family: Targets for therapeutic development against cancer and therapeutic strategies using monoclonal antibodies and tyrosine kinase inhibitors. Annu Rev Med 55:433–457
Misale S, Yaeger R, Hobor S et al (2012) Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486:532–536. https://doi.org/10.1038/nature11156
Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103:211–225
Neuzillet C, Hammel P, Tijeras-Raballand A et al (2013) Targeting the Ras-ERK pathway in pancreatic adenocarcinoma. Cancer Metastasis Rev 32:147–162
Montagut C, Settleman J (2009) Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett 283:125–134
Baier A, Szyszka R (2020) Compounds from natural sources as protein kinase inhibitors. Biomolecules 10:1–30
Pan MH, Lai CS, Wu JC, Ho CT (2011) Molecular mechanisms for chemoprevention of colorectal cancer by natural dietary compounds. Mol Nutr Food Res 55:32–45
Ye C, Li R, Xu L et al (2019) Effects of Baicalin on piglet monocytes involving PKC-MAPK signaling pathways induced by haemophilus parasuis. BMC Vet Res. https://doi.org/10.1186/s12917-019-1840-x
Awasthee N, Rai V, Chava S et al (2019) Targeting IκappaB kinases for cancer therapy. Semin Cancer Biol 56:12–24
Zhang HW, Hu JJ, Fu RQ et al (2018) Flavonoids inhibit cell proliferation and induce apoptosis and autophagy through downregulation of PI3Kγ mediated PI3K/AKT/mTOR/p70S6K/ULK signaling pathway in human breast cancer cells. Sci Rep. https://doi.org/10.1038/s41598-018-29308-7
Yin B, Fang DM, Zhou XL, Gao F (2019) Natural products as important tyrosine kinase inhibitors. Eur J Med Chem. 182
Jeong H, Phan ANH, Choi JW (2017) Anti-cancer effects of polyphenolic compounds in epidermal growth factor receptor tyrosine kinase inhibitor-resistant non-small cell lung cancer. Pharmacogn Mag 13:595–599. https://doi.org/10.4103/pm.pm_535_16
Yuan H, Ma Q, Cui H et al (2017) How can synergism of traditional medicines benefit from network pharmacology? Molecules 22:1135
Min X, Akella R, He H et al (2009) The structure of the MAP2K MEK6 reveals an autoinhibitory dimer. Struct (London, England : 1993) 17:96–104. https://doi.org/10.1016/j.str.2008.11.007
Borrelli N, Panebianco F, Condello V et al (2019) Characterization of activating mutations of the MEK1 Gene in papillary thyroid carcinomas. Thyroid : Offic J Am Thyroid Assoc 29:1279–1285. https://doi.org/10.1089/thy.2019.0065
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Broccatelli F, Salphati L, Plise E et al (2016) Predicting passive permeability of drug-like molecules from chemical structure: where are we? Mol Pharm 13:4199–4208. https://doi.org/10.1021/acs.molpharmaceut.6b00836
Press B, di Grandi D (2008) Permeability for intestinal absorption: caco-2 assay and related issues. Curr Drug Metab 9:893–900. https://doi.org/10.2174/138920008786485119
Al-Nour MY, Ibrahim MM, Elsaman T (2019) Ellagic acid, kaempferol, and quercetin from acacia nilotica: promising combined drug with multiple mechanisms of action. Current Pharmacol Reports 5:255–280. https://doi.org/10.1007/s40495-019-00181-w
Murugesan S, Kottekad S, Crasta I, et al (2021) Targeting COVID-19 (SARS-CoV-2) main protease through active phytocompounds of ayurvedic medicinal plants - Emblica officinalis (Amla), Phyllanthus niruri Linn. (Bhumi Amla) and Tinospora cordifolia (Giloy) - A molecular docking and simulation study. Computers in biology and medicine. 136: 104683. Doi: https://doi.org/10.1016/j.compbiomed.2021.104683
Tang D, Chen K, Huang L, Li J (2017) Pharmacokinetic properties and drug interactions of apigenin, a natural flavone. Expert Opinion Drug Metab Toxicol 13:323–330. https://doi.org/10.1080/17425255.2017.1251903
Seo H-S, Ku JM, Choi HS et al (2017) Apigenin overcomes drug resistance by blocking the signal transducer and activator of transcription 3 signaling in breast cancer cells. Oncol Rep 38:715–724. https://doi.org/10.3892/or.2017.5752
Conseil G, Baubichon-Cortay H, Dayan G et al (1998) Flavonoids: a class of modulators with bifunctional interactions at vicinal ATP- and steroid-binding sites on mouse P-glycoprotein. Proc Natl Acad Sci USA 95:9831–9836. https://doi.org/10.1073/pnas.95.17.9831
Oso BJ, Oyewo EB, Oladiji AT (2019) Influence of ethanolic extracts of dried fruit of Xylopia aethiopica (Dunal) A. Rich on haematological and biochemical parameters in healthy Wistar rats. Clin Phytosci 5:1–10. https://doi.org/10.1186/s40816-019-0104-4
Rheault TR, Caferro TR, Dickerson SH et al (2009) Thienopyrimidine-based dual EGFR/ErbB-2 inhibitors. Bioorg Med Chem Lett 19:817–820. https://doi.org/10.1016/j.bmcl.2008.12.011
Suzuki N, Shiota T, Watanabe F et al (2011) Synthesis and evaluation of novel pyrimidine-based dual EGFR/Her-2 inhibitors. Bioorg Med Chem Lett 21:1601–1606. https://doi.org/10.1016/j.bmcl.2011.01.119
Suzuki N, Shiota T, Watanabe F et al (2012) Discovery of novel 5-alkynyl-4-anilinopyrimidines as potent, orally active dual inhibitors of EGFR and Her-2 tyrosine kinases. Bioorg Med Chem Lett 22:456–460. https://doi.org/10.1016/j.bmcl.2011.10.103
Xu G, Searle LL, Hughes T, v, et al (2008) Discovery of novel 4-amino-6-arylaminopyrimidine-5-carbaldehyde oximes as dual inhibitors of EGFR and ErbB-2 protein tyrosine kinases. Bioorg Med Chem Lett 18:3495–3499. https://doi.org/10.1016/j.bmcl.2008.05.024
Li S, Guo C, Zhao H et al (2012) Synthesis and biological evaluation of 4-[3-chloro-4-(3-fluorobenzyloxy) anilino]-6-(3-substituted-phenoxy)pyrimidines as dual EGFR/ErbB-2 kinase inhibitors. Bioorg Med Chem 20:877–885. https://doi.org/10.1016/j.bmc.2011.11.056
Xu G, Abad MC, Connolly PJ et al (2008) 4-Amino-6-arylamino-pyrimidine-5-carbaldehyde hydrazones as potent ErbB-2/EGFR dual kinase inhibitors. Bioorg Med Chem Lett 18:4615–4619. https://doi.org/10.1016/j.bmcl.2008.07.020
Hughes T, v., Xu G, Wetter SK, et al (2008) A novel 5-[1,3,4-oxadiazol-2-yl]-N-aryl-4,6-pyrimidine diamine having dual EGFR/HER2 kinase activity: design, synthesis, and biological activity. Bioorg Med Chem Lett 18:4896–4899. https://doi.org/10.1016/j.bmcl.2008.07.057
Kawakita Y, Seto M, Ohashi T et al (2013) Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors. Bioorg Med Chem 21:2250–2261. https://doi.org/10.1016/j.bmc.2013.02.014
Tariq K, Ghias K (2016) Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol Med 13:120–135
Turanli B, Karagoz K, Gulfidan G et al (2019) A network-based cancer drug discovery: from integrated multi-omics approaches to precision medicine. Curr Pharm Des 24:3778–3790. https://doi.org/10.2174/1381612824666181106095959
Fan Q, Guo L, Guan J et al (2020) Network pharmacology-based study on the mechanism of Gegen Qinlian decoction against colorectal cancer. Evidence-based Complementary and Alternative Medicine. https://doi.org/10.1155/2020/8897879
Szklarczyk D, Santos A, von Mering C et al (2016) STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44:D380–D384. https://doi.org/10.1093/nar/gkv1277
Davies M, Nowotka M, Papadatos G et al (2015) ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res 43:W612–W620. https://doi.org/10.1093/nar/gkv352
Wang X, Shen Y, Wang S et al (2017) PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 45:W356–W360. https://doi.org/10.1093/nar/gkx374
Piñero J, Bravo Á, Queralt-Rosinach N et al (2017) DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 45:D833–D839. https://doi.org/10.1093/nar/gkw943
Chin CH, Chen SH, Wu HH et al (2014) cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. https://doi.org/10.1186/1752-0509-8-S4-S11
Lieu C, Kopetz S (2010) The Src family of protein tyrosine kinases: a new and promising target for colorectal cancer therapy. Clin Colorectal Cancer 9:89–94
Slattery ML, Mullany LE, Wolff RK et al (2019) The p53-signaling pathway and colorectal cancer: interactions between downstream p53 target genes and miRNAs. Genomics 111:762–771. https://doi.org/10.1016/j.ygeno.2018.05.006
Cheung LW, Mills GB (2016) Targeting therapeutic liabilities engendered by PIK3R1 mutations for cancer treatment. Pharmacogenomics 17:297–307
Wee P, Wang Z (2017) Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 9
Narayanankutty A (2019) PI3K/ Akt/ mTOR pathway as a therapeutic target for colorectal cancer: a review of preclinical and clinical evidence. Curr Drug Targets 20:1217–1226. https://doi.org/10.2174/1389450120666190618123846
Martinelli E, Morgillo F, Troiani T, Ciardiello F (2017) Cancer resistance to therapies against the EGFR-RAS-RAF pathway: the role of MEK. Cancer Treat Rev 53:61–69
Wagle N, Emery C, Berger MF et al (2011) Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 29:3085–3096. https://doi.org/10.1200/JCO.2010.33.2312
Pandurangan A, kumar, Divya T, Kumar K, et al (2018) Colorectal carcinogenesis: insights into the cell death and signal transduction pathways: a review. World Journal of Gastrointestinal Oncology 10:244–259
Wang X, Lin Y (2008) Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacol Sin 29:1275–1288
Press MF, Lenz H-J (2007) EGFR, HER2 and VEGF pathways: validated targets for cancer treatment. Drugs 67:2045–2075. https://doi.org/10.2165/00003495-200767140-00006
Folkman J (2007) Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discovery 6:273–286. https://doi.org/10.1038/nrd2115
J T, (2007) The role of VEGF and EGFR inhibition: implications for combining anti-VEGF and anti-EGFR agents. Molecular cancer research : MCR 5:203–220. https://doi.org/10.1158/1541-7786.MCR-06-0404
E S, A G, K G, S G, (2017) Plant flavone apigenin: an emerging anticancer agent. Current pharmacology reports 3:423–446. https://doi.org/10.1007/S40495-017-0113-2
H L, GO R, L L, et al (2009) Kaempferol inhibits angiogenesis and VEGF expression through both HIF dependent and independent pathways in human ovarian cancer cells. Nutr Cancer 61:554–563. https://doi.org/10.1080/01635580802666281
Sinha S, Patel S, Athar M et al (2019) Structure-based identification of novel sirtuin inhibitors against triple negative breast cancer: an in silico and in vitro study. Int J Biol Macromol 140:454–468. https://doi.org/10.1016/j.ijbiomac.2019.08.061
Parmar P, Rao P, Sharma A et al (2021) Meticulous assessment of natural compounds from NPASS database for identifying analogue of GRL0617, the only known inhibitor for SARS-CoV2 papain-like protease (PLpro) using rigorous computational workflow. Mol Diversity. https://doi.org/10.1007/s11030-021-10233-3
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10:449–461
Bowers KJ, Chow E, Xu H, et al (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, SC’06
Jorgensen WL, Tirado-Rives J (1988) The OPLS potential functions for proteins. energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 110:1657–1666. https://doi.org/10.1021/ja00214a001
Pires DEV, Blundell TL, Ascher DB (2015) pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58:4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Raudvere U, Kolberg L, Kuzmin I et al (2019) G:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47:W191–W198. https://doi.org/10.1093/nar/gkz369
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Sinha S, Khan S, Shukla S et al (2016) Cucurbitacin B inhibits breast cancer metastasis and angiogenesis through VEGF-mediated suppression of FAK/MMP-9 signaling axis. Int J Biochem Cell Biol 77:41–56. https://doi.org/10.1016/j.biocel.2016.05.014
Ribatti D (2008) R RE EV VI IE EW W The chick embryo chorioallantoic membrane in the study of tumor angiogenesis. Rom J Morphol Embryol 49:131–135
Demir R, Peros G, Hohenberger W (2011) Definition of the “Drug-Angiogenic-Activity-Index” that allows the quantification of the positive and negative angiogenic active drugs: a study based on the chorioallantoic membrane model. Pathol Oncol Res 17:309–313. https://doi.org/10.1007/S12253-010-9318-Y
Acknowledgements
The authors acknowledge our host institute, B.V. Patel Pharmaceutical Education Research, and Development (PERD) Centre for providing us with the facilities for our work. We also acknowledge the Department of chemistry, Gujarat University and Computational Chemistry Group (CCG@CUG), Central University of Gujarat, Gandhinagar, for providing computational resources.
Funding
This work was supported by the Department of Science & Technology (DST), Ministry of Science and Technology, Government of India for INSPIRE fellowship (Grant no. IF190211).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, A., Sinha, S., Rathaur, P. et al. Reckoning apigenin and kaempferol as a potential multi-targeted inhibitor of EGFR/HER2-MEK pathway of metastatic colorectal cancer identified using rigorous computational workflow. Mol Divers 26, 3337–3356 (2022). https://doi.org/10.1007/s11030-022-10396-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10396-7