Abstract
Anterior Gradient 2 (AGR2) has recently been reported as a tumor biomarker in various cancers, i.e., breast, prostate and lung cancer. Predominantly, AGR2 exists as a homodimer via a dimerization domain (E60-K64); after it is self-dimerized, it helps FGF2 and VEGF to homo-dimerize and promotes the angiogenesis and the invasion of vascular endothelial cells and fibroblasts. Up till now, no small molecule has been discovered to inhibit the AGR2–AGR2 homodimer. Therefore, the present study was performed to prepare a validated 3D structure of AGR2 by homology modeling and discover a small molecule by screening the FDA-approved drugs library on AGR2 homodimer as a target protein. Thirteen different homology models of AGR2 were generated based on different templates which were narrowed down to 5 quality models sorted by their overall Z-scores. The top homology model based on PDB ID = 3PH9 was selected having the best Z-score and was further assessed by Verify-3D, ERRAT and RAMPAGE analysis. Structure-based virtual screening narrowed down the large library of FDA-approved drugs to ten potential AGR2–AGR2 homodimer inhibitors having FRED score lower than − 7.8 kcal/mol in which the top 5 drugs’ binding stability was counter-validated by molecular dynamic simulation. To sum up, the present study prepared a validated 3D structure of AGR2 and, for the first time reported the discovery of 5 FDA-approved drugs to inhibit AGR2–AGR2 homodimer by using structure-based virtual screening. Moreover, the binding of the top 5 hits with AGR2 was also validated by molecular dynamic simulation.
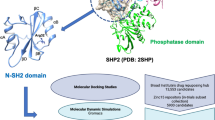
Graphic abstract
A validated 3D structure of Anterior Gradient 2 (AGR2) was prepared by homology modeling, which was used in virtual screening of FDA-approved drugs library for the discovery of prospective inhibitors of AGR2–AGR2 homodimer.






Similar content being viewed by others
References
Park SW, Zhen G, Verhaeghe C, Nakagami Y, Nguyenvu LT, Barczak AJ, Killeen N, Erle DJ (2009) The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc Natl Acad Sci 106:6950–6955. https://doi.org/10.1073/pnas.0808722106
Tiemann K, Garri C, Lee SB, Malihi PD, Park M, Alvarez RM, Yap LP, Mallick P, Katz JE, Gross ME (2019) Loss of ER retention motif of AGR2 can impact mTORC signaling and promote cancer metastasis. Oncogene 38:3003–3018. https://doi.org/10.1038/s41388-018-0638-9
Jia M, Guo Y, Zhu D, Zhang N, Li L, Jiang J, Dong Y, Xu Q, Zhang X, Wang M (2018) Pro-metastatic activity of AGR2 interrupts angiogenesis target bevacizumab efficiency via direct interaction with VEGFA and activation of NF-κB pathway. Biochim. Biophys. Acta. Mol Basis Dis 1864:1622–1633. https://doi.org/10.1016/j.bbadis.2018.01.021
Chevet E, Fessart D, Delom F, Mulot A, Vojtesek B, Hrstka R, Murray E, Gray T, Hupp T (2013) Emerging roles for the pro-oncogenic anterior gradient-2 in cancer development. Oncogene 32:2499–2509. https://doi.org/10.1038/onc.2012.346
Brychtova V, Vojtesek B, Hrstka R (2011) Anterior gradient 2: a novel player in tumor cell biology. Cancer lett 304:1–7. https://doi.org/10.1016/j.canlet.2010.12.023
Negi H, Merugu SB, Mangukiya HB, Li Z, Zhou B, Sehar Q, Kamle S, Mashausi DS, Wu Z, Li D (2019) Anterior Gradient-2 monoclonal antibody inhibits lung cancer growth and metastasis by upregulating p53 pathway and without exerting any toxicological effects: a preclinical study. Cancer Lett 449:125–134. https://doi.org/10.1016/j.canlet.2019.01.025
Patel P, Clarke C, Barraclough DL, Jowitt TA, Rudland PS, Barraclough R, Lian LY (2013) Metastasis-promoting anterior gradient 2 protein has a dimeric thioredoxin fold structure and a role in cell adhesion. J Mol Biol 425:929–943. https://doi.org/10.1016/j.jmb.2012.12.009
Higa A, Mulot A, Delom F, Bouchecareilh M, Nguyên DT, Boismenu D, Wise MJ, Chevet E (2011) Role of pro-oncogenic protein disulfide isomerase (PDI) family member anterior gradient 2 (AGR2) in the control of endoplasmic reticulum homeostasis. J Biol Chem 286:44855–44868. https://doi.org/10.1074/jbc.M111.275529
Guo H, Zhu Q, Yu X, Merugu SB, Mangukiya HB, Smith N, Li Z, Zhang B, Negi H, Rong R, Cheng K, Li D (2017) Tumor-secreted anterior gradient-2 binds to VEGF and FGF2 and enhances their activities by promoting their homodimerization. Oncogene 36:5098
Guo H, Chen H, Zhu Q, Yu X, Rong R, Merugu SB, Mangukiya HB, Li D (2016) A humanized monoclonal antibody targeting secreted anterior gradient 2 effectively inhibits the xenograft tumor growth. Biochem Biophys Res Commun 475:57–63. https://doi.org/10.1016/j.bbrc.2016.05.033
Garri C, Howell S, Tiemann K, Tiffany A, Jalali-Yazdi F, Alba MM, Katz JE, Takahashi TT, Landgraf R, Gross ME, Roberts RW (2018) Identification, characterization and application of a new peptide against anterior gradient homolog 2 (AGR2). Oncotarget 9:27363. https://doi.org/10.18632/oncotarget.25221
Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A (2019) Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18:41–58
Rani J, Silla Y, Borah K, Ramachandran S, Bajpai U (2019) Repurposing of FDA-approved drugs to target MurB and MurE enzymes in Mycobacterium tuberculosis. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2019.1637280
Dakshanamurthy S, Issa NT, Assefnia S, Seshasayee A, Peters OJ, Madhavan S, Uren A, Brown ML, Byers SW (2012) Predicting new indications for approved drugs using a proteochemometric method. J Med Chem 55:6832–6848. https://doi.org/10.1021/jm300576q
Khan SU, Ahemad N, Chuah L-H, Naidu R, Htar TT (2019) Sequential ligand-and structure-based virtual screening approach for the identification of potential G protein-coupled estrogen receptor-1 (GPER-1) modulators. RSC Adv 9:2525–2538. https://doi.org/10.1039/C8RA09318K
Gu X, Wang Y, Wang H, Wu H, Li W, Wang J, Li N (2020) Homology modeling, molecular dynamics and virtual screening of endothelin-A receptor for the treatment of pulmonary arterial hypertension. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1772106
Sun HP, Jiang ZY, Zhang MY, Lu MC, Yang TT, Pan Y, Huang HZ, Zhang XJ, You QD (2014) Novel protein–protein interaction inhibitor of Nrf2–Keap1 discovered by structure-based virtual screening. Med Chem Comm 5(1):93–98. https://doi.org/10.1039/C3MD00240C
Venselaar H, Joosten RP, Vroling B, Baakman C, Hekkelman ML, Krieger E, Vriend G (2010) Homology modelling and spectroscopy, a never-ending love story. Eur Biophys J 39:551–563. https://doi.org/10.1007/s00249-009-0531-0
Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins Struct Funct Bioinf 77:114–122. https://doi.org/10.1002/prot.22570
Krieger E, Koraimann G, Vriend G (2002) Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins Struct Funct Bioinf 47:393–402. https://doi.org/10.1002/prot.10104
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101(10):2525–2534. https://doi.org/10.1016/j.bpj.2011.10.024
Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model 50(4):572–584. https://doi.org/10.1021/ci100031x
McGann M (2011) FRED pose prediction and virtual screening accuracy. J Chem Inf Model 51(3):578–596. https://doi.org/10.1021/ci100436p
Khan A, Sayed SA, Khan MT, Saleem S, Ali A, Suleman M, Babar Z, Shafiq A, Khan M, Wei DQ (2020) Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro). J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1779128
Roe DR, Cheatham TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9(7):3084–3095. https://doi.org/10.1021/ct400341p
Consortium, Uniprot (2010) The universal protein resource (UniProt) in 2010. Nucleic Acids Res 38:142–148. https://doi.org/10.1093/nar/gkp846
Apweiler R, Bairoch A, Wu CH (2004) Protein sequence databases. Curr Opin Chem Biol 8:76–80. https://doi.org/10.1016/j.cbpa.2003.12.004
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zheng Z, Webb M, David JL (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292(2):195–202. https://doi.org/10.1006/jmbi.1999.3091
Xia J, Hu H, Xue W, Wang XS, Wu S (2018) The discovery of novel HDAC3 inhibitors via virtual screening and in vitro bioassay. J Enzyme Inhib Med Chem 33:525–535. https://doi.org/10.1080/14756366.2018.1437156
Acknowledgements
All the authors acknowledge the "OpenEye Scientific Software" for providing a free academic license to perform in silico studies. The principal author is grateful to Professor Huchen Zhou and Professor Dawei Li for deep insights into the project.
Funding
The lead author Shafi Ullah acknowledges China Scholarship Council (CSC) for funding this project.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest with anyone.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ullah, S., Khan, S.U., Khan, A. et al. Prospect of Anterior Gradient 2 homodimer inhibition via repurposing FDA-approved drugs using structure-based virtual screening. Mol Divers 26, 1399–1409 (2022). https://doi.org/10.1007/s11030-021-10263-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10263-x