Abstract
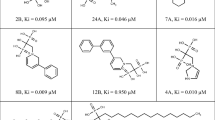
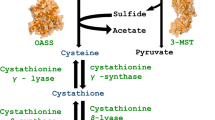
Leishmaniasis is one of the most neglected tropical diseases that demand immediate attention to the identification of new drug targets and effective drug candidates. The present study demonstrates the possibility of using threonine synthase (TS) as a putative drug target in leishmaniasis disease management. We report the construction of an effective homology model of the enzyme that appears to be structurally as well as functionally well conserved. The 200 nanosecond molecular dynamics data on TS with and without pyridoxal phosphate (PLP) shed light on mechanistic details of PLP-induced conformational changes. Moreover, we address some important structural and dynamic interactions in the PLP binding region of TS that are in good agreement with previously speculated crystallographic estimations. Additionally, after screening more than 44,000 compounds, we propose 10 putative inhibitor candidates for TS based on virtual screening data and refined Molecular Mechanics Generalized Born Surface Area calculations. We expect that structural and functional dynamics data disclosed in this study will help initiate experimental endeavors toward establishing TS as an effective antileishmanial drug target.
Graphic abstract









Similar content being viewed by others

Abbreviations
- ApoTS:
-
Apo form of leishmanial threonine synthase
- CHARMM:
-
Chemistry at Harvard Macromolecular Mechanics
- FDA:
-
Food and Drug Association
- GROMACS:
-
GROningen machine for chemical simulations
- HSK:
-
Homoserine kinase
- LGA:
-
Lamarckian genetic algorithm
- MD:
-
Molecular dynamics
- MM-GBSA:
-
Molecular Mechanics Generalized Born Surface Area
- MSA:
-
Multiple sequence alignment
- NAMD:
-
Nanoscale molecular dynamics
- NCBI:
-
National Center for Biotechnology Information
- NVE:
-
Ensemble-constant-energy, constant-volume, constant particle ensemble
- OPLS:
-
Optimized potential for liquid simulations
- PDB:
-
Protein data bank
- PDB-ID:
-
Protein data bank identifier
- pdbqt:
-
Protein data bank, partial charge (Q) and atom type (T)
- PHS:
-
Phospho-homoserine
- PLP:
-
Pyridoxal phosphate
- PME:
-
Particle mesh Ewald
- psf:
-
Protein structure file
- PyRX:
-
Python prescription
- RMSD:
-
Root-mean-square deviation
- RMSF:
-
Root-mean-square fluctuation
- SAVES server:
-
Structure analysis and verification server
- SPDBV:
-
Swiss PDB viewer
- SSB:
-
Sodium stibogluconate
- TIP3:
-
Three-site-transferrable intermolecular potential
- TS-PLP:
-
Leishmanial threonine synthase and pyridoxal phosphate complex
- UFF:
-
Universal force field
- VL:
-
Visceral leishmaniasis
- VMD:
-
Visual molecular dynamics
- VSGB:
-
Variable dielectric surface generalized born model
- WHO:
-
World Health Organization
- TS:
-
Threonine synthase
References
Youssefi MR, Moghaddas E, Tabari MA, Moghadamnia AA, Hosseini SM, Farash BRH, Ebrahimi MA, Mousavi NN, Fata A, Maggi F, Petrelli R, Dall’Acqua S, Benelli G, Sut S (2019) In vitro and in vivo effectiveness of carvacrol, thymol and linalool against Leishmania infantum. Molecules 24:2072. https://doi.org/10.3390/molecules24112072
Chakravarty J, Sundar S (2019) Current and emerging medications for the treatment of leishmaniasis. Expert Opin Pharmacother 7:1–15. https://doi.org/10.1080/14656566.2019.1609940
Tabbabi A (2019) Review of leishmaniasis in the Middle East and North Africa. Afr Health Sci 19:1329–1337. https://doi.org/10.4314/ahs.v19i1.4
Burza S, Croft SL, Boelaert M (2018) Leishmaniasis. Lancet 392:951–970. https://doi.org/10.1016/S0140-6736(18)31204-2
Braga SS (2019) Treating an old disease with new tricks: strategies based on host–guest chemistry for leishmaniasis therapy. J Incl Phenom Macrocycl Chem 93:145–155. https://doi.org/10.1007/s10847-019-00885-y
WHO Leishmaniasis. http://www.who.int/leishmaniasis/en/. Accessed 4 May 2019
Bruschi F, Gradoni L (2018) The leishmaniases: old neglected tropical diseases. Springer International, Berlin. https://doi.org/10.1007/978-3-319-72386-0
Hirve S, Boelaert M, Matlashewski G, Mondal D, Arana B, Kroeger A, Olliaro P (2016) Transmission Dynamics of visceral leishmaniasis in the Indian subcontinent—a systematic literature review. PLoS Negl Trop Dis 10:e0004896. https://doi.org/10.1016/S0140-6736(18)31204-2.e0004896
Ghorbani M, Farhoudi R (2017) Leishmaniasis in humans: drug or vaccine therapy? Drug Des Devel Ther 12:25–40. https://doi.org/10.2147/DDDT.S146521
Alves F, Bilbe G, Blesson S, Goyal V, Monnerat S, Mowbray C, Muthoni Ouattara G, Pécoul B, Rijal S, Rode J, Solomos A, Strub-Wourgaft N, Wasunna M, Wells S, Zijlstra EE, Arana B, Alvar J (2018) Recent development of visceral leishmaniasis treatments: successes, pitfalls, and perspectives. Clin Microbiol Rev. https://doi.org/10.1128/CMR.00048-18
Ponte-Sucre A, Gamarro F, Dujardin JC, Barrett MP, López-Vélez R, García-Hernández R, Pountain AW, Mwenechanya R, Papadopoulou B (2017) Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0006052.e0006052
Chakravarty J, Sundar S (2010) Drug resistance in leishmaniasis. J Glob Infect Dis 2:167–176. https://doi.org/10.4103/0974-777X.62887
Rijal S, Chappuis F, Singh R, Bovier PA, Acharya P, Karki BM, Das ML, Desjeux P, Loutan L, Koirala S (2003) Treatment of visceral leishmaniasis in south-eastern Nepal: decreasing efficacy of sodium stibogluconate and need for a policy to limit further decline. Trans R Soc Trop Med Hyg 97:350–354. https://doi.org/10.1016/S0035-9203(03)90167-2
Sundar S, More DK, Singh MK, Singh VP, Sharma S, Makharia A, Kumar PC, Murray HW (2000) Failure of pentavalent antimony in visceral leishmaniasis in India: report from the center of the Indian epidemic. Clin Infect Dis 31:1104–1107. https://doi.org/10.1086/318121
Sundar S, Jha TK, Thakur C, Engel J, Sindermann H, Fischer C, Junge K, Bryceson A, Berman J (2002) Oral miltefosine for Indian visceral leishmaniasis. N Engl J Med 347:1739–1746. https://doi.org/10.1056/NEJMoa021556
Singh OP, Singh B, Chakravarty J, Sundar S (2016) Current challenges in treatment options for visceral leishmaniasis in India: a public health perspective. Infect Dis Poverty 5:19. https://doi.org/10.1186/s40249-016-0112-2
Sundar S, Singh A (2016) Recent developments and future prospects in the treatment of visceral leishmaniasis. Ther Adv Infect Dis 3:98–109. https://doi.org/10.1177/2049936116646063
Maintz EM, Hassan M, Huda MM, Ghosh D, Hossain MS, Alim A, Kroeger A, Arana B, Mondal D (2014) Introducing single dose liposomal amphotericin B for the treatment of visceral leishmaniasis in rural bangladesh: feasibility and acceptance to patients and health staff. J Trop Med 2014:676817. https://doi.org/10.1155/2014/676817
Pountain AW, Weidt SK, Regnault C, Bates PA, Donachie AM, Dickens NJ, Barrett MP (2019) Genomic instability at the locus of sterol C24-methyltransferase promotes amphotericin B resistance in Leishmania parasites. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0007052.e0007052
Purkait B, Kumar A, Nandi N, Sardar AH, Das S, Kumar S, Pandey K, Ravidas V, Kumar M, De T, Singh D, Das P (2012) Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrob Agents Chemother 56:1031–1041. https://doi.org/10.1128/AAC.00030-11
Srivastava P, Dayama A, Mehrotra S, Sundar S (2011) Diagnosis of visceral leishmaniasis. Trans R Soc Trop Med Hyg 105:1–6. https://doi.org/10.1016/j.trstmh.2010.09.006
Agrawal VK, Singh Z (2006) Miltefosine: first oral drug for treatment of visceral leishmaniasis. Med J Armed Forces India 62:66–67. https://doi.org/10.1016/S0377-1237(06)80162-0
Pijpers J, den Boer ML, Essink DR, Ritmeijer K (2019) The safety and efficacy of miltefosine in the long-term treatment of post-kala-azar dermal leishmaniasis in South Asia—a review and meta-analysis. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0007173.e0007173
Tiwari N, Kumar A, Singh A, Bajpai S, Agrahari AK, Kishore D, Tiwari VK, Singh RK (2019) Leishmaniasis control: limitations of current drugs and prospects of natural products. In: Brahmachari G (ed) Discovery and development of therapeutics from natural products against neglected tropical diseases, 1st edn. Elsevier, Amsterdam, pp 293–350. https://doi.org/10.1016/B978-0-12-815723-7.00008-0
Contreras MEM (2019) Chemotherapy used in the treatment of visceral leishmaniasis. CPQ Microbiol 3:1–14
Ramesh V, Kaushal H, Mishra AK, Singh R, Salotra P (2015) Clinico-epidemiological analysis of Post kala-azar dermal leishmaniasis (PKDL) cases in India over last two decades: a hospital based retrospective study. BMC Public Health 15:1092. https://doi.org/10.1186/s12889-015-2424-8
Mondelaers A, Sanchez-Cañete MP, Hendrickx S, Eberhardt E, Garcia-Hernandez R, Lachaud L, Cotton J, Sanders M, Cuypers B, Imamura H, Dujardin JC, Delputte P, Cos P, Caljon G, Gamarro F, Castanys S, Maes L (2016) Genomic and molecular characterization of miltefosine resistance in Leishmania infantum strains with either natural or acquired resistance through experimental selection of intracellular amastigotes. PLoS ONE. https://doi.org/10.1371/journal.pone.0154101.e0154101
Vacchina P, Norris-Mullins B, Abengózar MA, Viamontes CG, Sarro J, Stephens MT, Pfrender ME, Rivas L, Morales MA (2016) Genomic appraisal of the multifactorial basis for in vitro acquisition of miltefosine resistance in Leishmania donovani. Antimicrob Agents Chemother 60:4089–4100. https://doi.org/10.1128/AAC.00478-16
Kulshrestha A, Sharma V, Singh R, Salotra P (2014) Comparative transcript expression analysis of miltefosine-sensitive and miltefosine-resistant Leishmania donovani. Parasitol Res 113:1171–1184. https://doi.org/10.1007/s00436-014-3755-6
Srivastava S, Mishra J, Gupta AK, Singh A, Shankar P, Singh S (2017) Laboratory confirmed miltefosine resistant cases of visceral leishmaniasis from India. Parasit Vectors 10:49. https://doi.org/10.1186/s13071-017-1969-z
Meshram RJ, Goundge MB, Kolte BS, Gacche RN (2019) An in silico approach in identification of drug targets in Leishmania: a subtractive genomic and metabolic simulation analysis. Parasitol Int 69:59–70. https://doi.org/10.1016/j.parint.2018.11.006
Alexander FW, Sandmeier E, Mehta PK, Christen P (1994) Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Eur J Biochem 219:953–960. https://doi.org/10.1111/j.1432-1033.1994.tb18577.x
Grishin NV, Phillips MA, Goldsmith EJ (1995) Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci 4:1291–1304. https://doi.org/10.1002/pro.5560040705
Mehta PK, Christen P (2000) The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv Enzymol Relat Areas Mol Biol 74:129–184. https://doi.org/10.1002/9780470123201.ch4
Garrido-Franco M, Ehlert S, Messerschmidt A, Marinkovic S, Huber R, Laber B, Bourenkov GP, Clausen T (2002) Structure and function of threonine synthase from yeast. J Biol Chem 277:12396–12405. https://doi.org/10.1074/jbc.M108734200
Flavin M, Slaughter C (1960) Purification and properties of threonine synthetase of Neurospora. J Biol Chem 235:1103–1108
Skarstedt MT, Greer SB (1973) Threonine synthetase of Bacillus subtilis. The nature of an associated dehydratase activity. J Biol Chem 248:1032–1044
Parsot C, Cossart P, Saint-Girons I, Cohen GN (1983) Nucleotide sequence of thrC and of the transcription termination region of the threonine operon in Escherichia coli K12. Nucleic Acids Res 11:7331–7345. https://doi.org/10.1093/nar/11.21.7331
Shames SL, Ash DE, Wedler FC, Villafranca JJ (1984) Interaction of aspartate and aspartate-derived antimetabolites with the enzymes of the threonine biosynthetic pathway of Escherichia coli. J Biol Chem 259:15331–15339
Shames SL, Wedler FC (1984) Homoserine kinase of Escherichia coli: kinetic mechanism and inhibition by l-aspartate semialdehyde. Arch Biochem Biophys 235:359–370. https://doi.org/10.1016/0003-9861(84)90209-1
Laber B, Gerbling KP, Harde C, Neff KH, Nordhoff E, Pohlenz HD (1994) Mechanisms of interaction of Escherichia coli threonine synthase with substrates and inhibitors. Biochemistry 33:3413–3423. https://doi.org/10.1021/bi00177a035
Laber B, Maurer W, Hanke C, Gräfe S, Ehlert S, Messerschmidt A, Clausen T (1999) Characterization of recombinant Arabidopsis thaliana threonine synthase. Eur J Biochem 263:212–221. https://doi.org/10.1046/j.1432-1327.1999.00487.x
Curien G, Dumas R, Ravanel S, Douce R (1996) Characterization of an Arabidopsis thaliana cDNA encoding an S-adenosylmethionine-sensitive threonine synthase. Threonine synthase from higher plants. FEBS Lett 390:85–90. https://doi.org/10.1016/0014-5793(96)00633-3
Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31:365–370. https://doi.org/10.1093/nar/gkg095
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Westbrook J, Feng Z, Chen L, Yang H, Berman HM (2003) The protein data bank and structural genomics. Nucleic Acids Res 31:489–491. https://doi.org/10.1093/nar/gkg068
Meshram RJ, Gavhane AJ, Gaikar RB, Bansode TS, Maskar AU, Gupta AK, Sohni SK, Patidar MA, Pandey TR, Jangle SN (2010) Sequence analysis and homology modeling of laccase from Pycnoporus cinnabarinus. Bioinformation 5:150–154. https://doi.org/10.6026/97320630005150
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. https://doi.org/10.1006/jmbi.1993.1626
Madhusudhan MS, Marti-Renom MA, Sanchez R, Sali A (2006) Variable gap penalty for protein sequence-structure alignment. Protein Eng Des Sel 19:129–133. https://doi.org/10.1093/protein/gzj005
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453. https://doi.org/10.1016/0022-2836(70)90057-4
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. https://doi.org/10.1002/elps.1150181505
Van Gunsteren WF, Brunne RM, Gros P, van Schaik RC, Schiffer CA, Torda AE (1994) Accounting for molecular mobility in structure determination based on nuclear magnetic resonance spectroscopic and X-ray diffraction data. Methods Enzymol 239:619–654. https://doi.org/10.1016/s0076-6879(94)39024-x
Hooft RW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381:272. https://doi.org/10.1038/381272a0
Laskowski RA, MacAurther MW, Moss DS, Thornton JM (1993) PROCHECK—a program to check the stereochemical quality of proteins. J Appl Cryst 26:47–60. https://doi.org/10.1006/jmbi.1996.0663
Castrignanò T, De Meo PD, Cozzetto D, Talamo IG, Tramontano A (2006) The PMDB protein model database. Nucleic Acids Res 34:D306–D309. https://doi.org/10.1093/nar/gkj105
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152. https://doi.org/10.1002/jcc.21256
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Ambhore AN, Kamble SS, Kadam SN, Kamble RD, Hebade MJ, Hese SV, Gaikwad MV, Meshram RJ, Gacche RN, Dawane BS (2019) Design, synthesis and in silico study of pyridine based 1,3,4-oxadiazole embedded hydrazinecarbothioamide derivatives as potent anti-tubercular agent. Comput Biol Chem 80:54–65. https://doi.org/10.1016/j.compbiolchem.2019.03.002
Patil KK, Meshram RJ, Barage SH, Gacche RN (2019) Dietary flavonoids inhibit the glycation of lens proteins: implications in the management of diabetic cataract. 3 Biotech 9:47. https://doi.org/10.1007/s13205-019-1581-3
Kolte BS, Londhe SR, Solanki BR, Gacche RN, Meshram RJ (2018) FilTer BaSe: a web accessible chemical database for small compound libraries. J Mol Graph Model 80:95–103. https://doi.org/10.1016/j.jmgm.2017.12.020
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminform 3:33. https://doi.org/10.1186/1758-2946-3-33
Azizian H, Bahrami H, Pasalar P, Amanlou M (2010) Molecular modeling of Helicobacter pylori arginase and the inhibitor coordination interactions. J Mol Graph Model 28:626–635. https://doi.org/10.1016/j.jmgm.2009.12.007
Dassault Systèmes BIOVIA, Discovery Studio Visualizer, v 17.2.0.16349, San Diego: Dassault Systèmes (2016)
Cosconati S, Forli S, Perryman AL, Harris R, Goodsell DS, Olson AJ (2010) Virtual screening with autodock: theory and practice. Expert Opin Drug Discov 5:597–607. https://doi.org/10.1517/17460441.2010.484460
Schrödinger Release 2019-1: Prime v3.2, Schrödinger, LLC, New York (2019)
Du J, Sun H, Xi L, Li J, Yang Y, Liu H, Yao X (2011) Molecular modeling study of checkpoint kinase 1 inhibitors by multiple docking strategies and prime/MM-GBSA calculation. J Comput Chem 32:2800–2809. https://doi.org/10.1002/jcc.21859
Patil A, Duggal H, Bagul KT, Kamble S, Lokhande P, Gacche R, Meshram R (2020) Synthesis of new 3-arylaminophthalides and 3-indolyl-phthalides using ammonium chloride, evaluation of their anti-mycobacterial potential and docking study. Comb Chem High Throughput Screen. https://doi.org/10.2174/1386207323666200422082754
Meshram RJ, Bagul KT, Pawnikar SP, Barage SH, Kolte BS, Gacche RN (2020) Known compounds and new lessons: structural and electronic basis of flavonoid-based bioactivities. J Biomol Struct Dyn 38:1168–1184. https://doi.org/10.1080/07391102.2019.1597770
Humphrey W, Dalke A, Schulten KK (1995) VMD—visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. https://doi.org/10.1002/jcc.20289
Yesselman JD, Price DJ, Knight JL, Brooks CL (2012) MATCH: an atom-typing toolset for molecular mechanics force fields. J Comput Chem 33:189–202. https://doi.org/10.1002/jcc.21963
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Ryckaert JP, Ciccotti G, Berendsen JCH (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an Nlog·(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Hoover WG (1985) Canonical dynamics: equilibrium phase-space distributions. Phys Rev A Gen Phys 31:1695–1697. https://doi.org/10.1103/physreva.31.1695
Feller SE, Zhang Y, Pastor RW (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103:4613–4621. https://doi.org/10.1063/1.470648
Martyna GJ, Tobias DJ, Klein ML (1994) Constant pressure molecular dynamics algorithms. J Chem Phys 101:4177–4189. https://doi.org/10.1063/1.467468
Drug Design Laboratory—VEGA ZZ. https://nova.disfarm.unimi.it/cms/index.php?Software_projects:VEGA_ZZ. Accessed 4 May 2019
Daura X, Gademann K, Jaun B, Seebach D, VanGunsteren WF, Mark AE (1999) Peptide folding: when simulation meets experiment. Angew Chem Int Ed 38:236–240. https://doi.org/10.1002/(SICI)1521-3773(19990115)38:1/23.3.CO;2-D
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. https://doi.org/10.1021/ci200227u
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134. https://doi.org/10.1093/protein/8.2.127
Knapp B, Lederer N, Omasits U, Schreiner W (2010) vmdICE: a plug-in for rapid evaluation of molecular dynamics simulations using VMD. J Comput Chem 31:2868–2873. https://doi.org/10.1002/jcc.21581
Maskar AU, Meshram RJ (2013) Homology modeling of chorismate synthase from Brucella melitensis: a novel target molecule. Res Rev J Microbiol Biotechnol 2:7–18
Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS (2003) VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res 31:3316–3319. https://doi.org/10.1093/nar/gkg565
Bender DA (1985) Transaminases. In: Christen P, Metzler DE (eds) Wiley-Interscience, New York, p 643. https://doi.org/10.1016/0014-5793(85)80462-2
Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR (1988) Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem 263:17857–17871
Burkhard P, Rao GS, Hohenester E, Schnackerz KD, Cook PF, Jansonius JN (1998) Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J Mol Biol 283:121–133. https://doi.org/10.1006/jmbi.1998.2037
Jansonius JN (1998) Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol 8:759–769. https://doi.org/10.1016/S0959-440X(98)80096-1
Voet D, Voet JG, Pratt CW (2006) Fundamentals of biochemistry, 2nd edn. Wiley, New York
Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry, 5th edn. W. H. Freeman, New York
Anjana R, Vaishnavi MK, Sherlin D, Kumar SP, Naveen K, Kanth PS, Sekar K (2012) Aromatic-aromatic interactions in structures of proteins and protein-DNA complexes: a study based on orientation and distance. Bioinformation 8:1220–1224. https://doi.org/10.6026/97320630081220
Loyola PKR, Campos-Rodríguez R, Bello M, Rojas-Hernández S, Zimic M, Quiliano M, Briz V, Muñoz-Frenández MA, Tolentino-López L, Correa-Basurto J (2013) Theoretical analysis of the neuraminidase epitope of the Mexican A H1N1 influenza strain, and experimental studies on its interaction with rabbit and human hosts. Immunol Res 56:44–60. https://doi.org/10.1007/s12026-013-8385-z
Prati F, Goldman-Pinkovich A, Lizzi F, Belluti F, Koren R, Zilberstein D, Bolognesi ML (2013) Quinone-amino acid conjugates targeting leishmania amino acid transporters. PLoS ONE. https://doi.org/10.1371/journal.pone.0107994
Marchese L, Nascimento JF, Damasceno FS, Bringaud F, Michels PAM, Silber AM (2018) The uptake and metabolism of amino acids, and their unique role in the biology of pathogenic trypanosomatids. Pathogens 7:36. https://doi.org/10.3390/pathogens7020036
Ter Kuile BH, Opperdoes FR (1992) A chemostat study on proline uptake and metabolism of Leishmania donovani. J Protozool 39:555–558. https://doi.org/10.1111/j.1550-7408.1992.tb04850.x
Mukkada AJ, Simon MW (1977) Leishmania tropica: uptake of methionine by promastigotes. Exp Parasitol 42:87–96. https://doi.org/10.1016/0014-4894(77)90065-0
Simon MW, Mukkada AJ (1977) Leishmania tropica: regulation and specificity of the methionine transport system in promastigotes. Exp Parasitol 42:97–105. https://doi.org/10.1016/0014-4894(77)90066-2
Bonay P, Cohen BE (1983) Neutral amino acid transport in Leishmania promastigotes. Biochim Biophys Acta 731:222–228. https://doi.org/10.1016/0005-2736(83)90012-3
Paes LS, Galvez Rojas RL, Daliry A, Floeter-Winter LM, Ramirez MI, Silber AM (2008) Active transport of glutamate in Leishmania (Leishmania) amazonensis. J Eukaryot Microbiol 55:382–387. https://doi.org/10.1111/j.1550-7408.2008.00346.x
Kandpal M, Fouce RB, Pal A, Guru PY, Tekwani BL (1995) Kinetics and molecular characteristics of arginine transport by Leishmania donovani promastigotes. Mol Biochem Parasitol 71:193–201. https://doi.org/10.1016/0166-6851(95)00042-Y
Shaked-Mishan P, Suter-Grotemeyer M, Yoel-Almagor T, Holland N, Zilberstein D, Rentsch D (2006) A novel high-affinity arginine transporter from the human parasitic protozoan Leishmania donovani. Mol Microbiol 60:30–38. https://doi.org/10.1111/j.1365-2958.2006.05060.x
Darlyuk I, Goldman A, Roberts SC, Ullman B, Rentsch D, Zilberstein D (2009) Arginine homeostasis and transport in the human pathogen Leishmania donovani. J Biol Chem 284:19800–19807. https://doi.org/10.1074/jbc.M901066200
Castilho-Martins EA, Laranjeira da Silva MF, dos Santos MG, Muxel SM, Floeter-Winter LM (2011) Axenic Leishmania amazonensis promastigotes sense both the external and internal arginine pool distinctly regulating the two transporter-coding genes. PLoS ONE. https://doi.org/10.1371/journal.pone.0027818.e27818
Aoki JI, Muxel SM, Zampieri RA, Acuña SM, Fernandes JCR, Vanderlinde RH, Sales MCOP, Floeter-Winter LM (2017) L-arginine availability and arginase activity: characterization of amino acid permease 3 in Leishmania amazonensis. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0006025.e0006025
Inbar E, Canepa GE, Carrillo C, Glaser F, Suter Grotemeyer M, Rentsch D, Zilberstein D, Pereira CA (2012) Lysine transporters in human trypanosomatid pathogens. Amino Acids 42:347–360. https://doi.org/10.1007/s00726-010-0812-z
dos Santos MG, Paes LS, Zampieri RA, da Silva MF, Silber AM, Floeter-Winter LM (2009) Biochemical characterization of serine transport in Leishmania (Leishmania) amazonensis. Mol Biochem Parasitol 163:107–113. https://doi.org/10.1016/j.molbiopara.2008.11.001
Duszenko M, Ferguson MA, Lamont GS, Rifkin MR, Cross GA (1985) Cysteine eliminates the feeder cell requirement for cultivation of Trypanosoma brucei bloodstream forms in vitro. J Exp Med 162:1256–1263. https://doi.org/10.1084/jem.162.4.1256
Canepa GE, Bouvier LA, Miranda MR, Uttaro AD, Pereira CA (2009) Characterization of Trypanosoma cruzi l-cysteine transport mechanisms and their adaptive regulation. FEMS Microbiol Lett 292:27–32. https://doi.org/10.1111/j.1574-6968.2008.01467.x
Barisón MJ, Damasceno FS, Mantilla BS, Silber AM (2016) The active transport of histidine and its role in ATP production in Trypanosoma cruzi. J Bioenergy Biomembr 48:437–449. https://doi.org/10.1007/s10863-016-9665-9
Damasceno FS, Barisón MJ, Crispim M, Souza ROO, Marchese L, Silber AM (2018) l-Glutamine uptake is developmentally regulated and is involved in metacyclogenesis in Trypanosoma cruzi. Mol Biochem Parasitol 224:17–25. https://doi.org/10.1016/j.molbiopara.2018.07.007
Manchola NC, Rapado LN, Barisón MJ, Silber AM (2016) Biochemical characterization of branched chain amino acids uptake in Trypanosoma cruzi. J Eukaryot Microbiol 63:299–308. https://doi.org/10.1111/jeu.12278
Canepa GE, Bouvier LA, Urias U, Miranda MR, Colli W, Alves MJ, Pereira CA (2005) Aspartate transport and metabolism in the protozoan parasite Trypanosoma cruzi. FEMS Microbiol Lett 247:65–71. https://doi.org/10.1016/j.femsle.2005.04.029
Fricker SP, Jones SE, Ellory JC, Angus JM, Klein RA (1984) Threonine uptake in Trypanosoma brucei. Mol Biochem Parasitol 11:215–223. https://doi.org/10.1016/0166-6851(84)90067-7
Acknowledgements
We sincerely thank Dr. Sangeeta Sawant, Director, Bioinformatics Centre, Savitribai Phule Pune University, Pune, for providing infrastructure and support throughout the project. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (MP4 18138 kb)
Rights and permissions
About this article
Cite this article
Meshram, R.J., Bagul, K.T., Aouti, S.U. et al. Modeling and simulation study to identify threonine synthase as possible drug target in Leishmania major. Mol Divers 25, 1679–1700 (2021). https://doi.org/10.1007/s11030-020-10129-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-020-10129-8