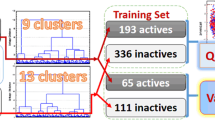
Abstract
Selective inhibition of Bcl-2 and Bcl-xL proteins due to their dual inhibition toxicity plays an important role in treatment of cancer and chemotherapy effectiveness; therefore, in the last decade, discovery of selective inhibitors for Bcl-2 and Bcl-xL proteins has become a significant and important research topic. The present contribution paves the way for characterization of molecular features which induce selectivity for inhibition of Bcl-2 and Bcl-xL. In this line, a total of 1534 molecules related to inhibition of Bcl-2 and Bcl-xL proteins were collected from Binding Database. A diverse set of molecular descriptors was calculated for each molecule, and the best subset of descriptors were selected using variable importance in projection (VIP) approach. The molecules were classified according to their therapeutic targets (Bcl-2/Bcl-xL) and activities. Partial least square-discriminate analysis (PLS-DA) and supervised Kohonen network (SKN) models were utilized to relate the molecular structures of chemicals to their activities and selectivities. According to the VIP-selected descriptors physicochemical properties, such as polarity number, number of branches, size and cyclicity of the molecule, flexibility, functional counts and constitutional descriptors, all affect the activities of Bcl-2 and Bcl-xL inhibitors. The performances of PLS-DA and SKN methods were evaluated based on statistical parameters derived from the confusion matrices. The models were validated using tenfold cross-validation and an external test set. The best statistical results were obtained by implementing the SKN model. The classification rates range from 93.5 to 79.1% for the training and validation procedure for the optimized SKN models. The high values of the obtained classification rates demonstrate that the information provided in this work would be useful to design new drugs with selective inhibitory activities toward Bcl-2 or Bcl-xL proteins for more effective treatment of cancer.
Graphical abstract











Similar content being viewed by others
References
World Health Organization (2017) WHO’s work on cancer. http://www.who.int/cancer/en/. Accessed 23 Aug 2017
Ashkenazi A, Fairbrother WJ, Leverson JD, Souers AJ (2017) From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat Rev Drug Discov 16:273–284. https://doi.org/10.1182/blood-2011-03-344812
Wickman G, Julian L, Olson MF (2012) How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ 19(5):735–742. https://doi.org/10.1038/cdd.2012.25
Vogler M, Hamali HA, Sun XM, Bampton ET, Dinsdale D, Snowden RT, Dyer MJ, Goodall AH, Cohen GM (2011) BCL2/BCL-XL inhibition induces apoptosis, disrupts cellular calcium homeostasis, and prevents platelet activation. Blood 117(26):7145–7154. https://doi.org/10.1038/nrd.2016.253
Zhao DP, Ding XW, Peng JP, Zheng YX, Zhang SZ (2005) Prognostic significance of bcl-2 and p53 expression in colorectal carcinoma. J Zhejiang Univ Sci B 6:1163–1169. https://doi.org/10.1631/jzus.2005.B1163
Correia C, Schneider PA, Dai H, Dogan A, Maurer MJ, Church AK, Novak AJ, Feldman AL, Wu X, Ding H, Meng XW (2015) BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood 125:658–667. https://doi.org/10.1182/blood-2014-04-571786
Merino D, Lok SW, Visvader JE, Lindeman GJ (2016) Targeting BCL-2 to enhance vulnerability to therapy in estrogen receptor-positive breast cancer. Oncogene 35:1877–1887. https://doi.org/10.1038/onc.2015.287
Enyedy IJ, Ling Y, Nacro K, Tomita Y, Wu X, Cao Y, Guo R, Li B, Zhu X, Huang Y, Long YQ, Roller PP, Yang D, Wang S (2001) Discovery of small-molecule inhibitors of Bcl-2 through structure-based computer screening. J Med Chem 44:4313–4324. https://doi.org/10.1021/jm010016f
Colak S, Zimberlin CD, Fessler E, Hogdal L, Prasetyanti PR, Grandela CM, Letai A, Medema JP (2014) Decreased mitochondrial priming determines chemoresistance of colon cancer stem cells. Cell Death Differ 21:1170–1177. https://doi.org/10.1038/cdd.2014.37
Zeuner A, Francescangeli F, Contavalli P, Zapparelli G, Apuzzo T, Eramo A, Baiocchi M, De Angelis ML, Biffoni M, Sette G, Todaro M, Stassi G, Maria RD (2014) Elimination of quiescent/slow-proliferating cancer stem cells by Bcl-XL inhibition in non-small cell lung cancer. Cell Death Differ 21:1877–1888. https://doi.org/10.1038/cdd.2014.105
Wong M, Tan N, Zha J, Peale FV, Yue P, Fairbrother WJ, Belmont LD (2012) Navitoclax (ABT-263) reduces Bcl-x(L)-mediated chemoresistance in ovarian cancer models. Mol Cancer Ther 11:1026–1035. https://doi.org/10.1158/1535-7163.MCT-11-0693
Ikezawa K, Hikita H, Shigekawa M, Iwahashi K, Eguchi H, Sakamori R, Tatsumi T, Takehara T (2017) Increased Bcl-xL expression in pancreatic neoplasia promotes carcinogenesis by inhibiting senescence and apoptosis. Cell Mol Gastroenterol Hepatol 4:185–200. https://doi.org/10.1016/j.jcmgh.2017.02.001
Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DC et al (2013) ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19:202–209. https://doi.org/10.1038/nm.3048
Wang JL, Liu D, Zhang ZJ, Shan S, Han X, Srinivasula SM, Croce CM, Alnemri ES, Huang Z (2000) Structure-based discovery of an organic compound that binds Bcl-2 protein and induces apoptosis of tumor cells. Proc Natl Acad Sci India A 97(13):7124–7129. https://doi.org/10.1073/pnas.97.13.7124
Enyedy IJ, Ling Y, Nacro K, Tomita Y, Wu X, Cao Y, Guo R, Li B, Zhu X, Huang Y, Long YQ (2001) Discovery of small-molecule inhibitors of Bcl-2 through structure-based computer screening. J Med Chem 44(25):4313–4324. https://doi.org/10.1021/jm010016f
Kanakaveti V, Sakthivel R, Rayala SK, Gromiha MM (2017) Importance of functional groups in predicting the activity of small molecule inhibitors for Bcl-2 and Bcl-xL. Chem Biol Drug Des 90(2):308–316. https://doi.org/10.1111/cbdd.12952
Mukherjee P, Desai P, Zhou YD, Avery M (2010) Targeting the BH3 domain mediated protein–protein interaction of Bcl-xL through virtual screening. J Chem Inf Model 50(5):906–923. https://doi.org/10.1021/ci1000373
Lama D, Sankararamakrishnan R (2008) Anti‐apoptotic Bcl‐xL protein in complex with BH3 peptides of pro‐apoptotic Bak, Bad, and Bim proteins: comparative molecular dynamics simulations. Proteins Struct Funct Bioinform 73(2):492–514. https://doi.org/10.1002/prot.22075
Wakui N, Yoshino R, Yasuo N, Ohue M, Sekijima M (2018) Exploring the selectivity of inhibitor complexes with Bcl-2 and Bcl-xL: a molecular dynamics simulation approach. J Mol Graph Model 79:166–174. https://doi.org/10.1016/j.jmgm.2017.11.011
Almerico AM, Tutone M, Lauria A (2009) In-silico screening of new potential Bcl-2/Bcl-xL inhibitors as apoptosis modulators. J Mol Model 15(4):349–355. https://doi.org/10.1007/s00894-008-0405-x
Aboalhaija NH, Zihlif MA, Taha MO (2016) Discovery of new selective cytotoxic agents against Bcl-2 expressing cancer cells using ligand-based modeling. Chem Biol Interact 250:12–26. https://doi.org/10.1016/j.cbi.2016.03.006
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J (2016) Binding DB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44:D0145–D1053. https://doi.org/10.1093/nar/gkv1072
Jalali-Heravi M, Mani-Varnosfaderani A, Jahromi PE, Mahmoodi MM, Taherinia D (2011) Classification of anti-HIV compounds using counter propagation artificial neural networks and decision trees. SAR QSAR Environ Res 22(7–8):639–660. https://doi.org/10.1080/1062936X.2011.623318
Jalali-Heravi M, Mani-Varnosfaderani A, Valadkhani A (2013) Integrated one-against-one classifiers as tools for virtual screening of compound databases: a case study with CNS inhibitors. Mol Inf 32:742–753. https://doi.org/10.1002/minf.201200126
Jalali-Heravi M, Mani-Varnosfaderani A (2012) Navigating drug-like chemical space of anticancer molecules using genetic algorithms and counter propagation artificial neural networks. Mol Inf 31:63–74. https://doi.org/10.1002/minf.201100098
Neiband MS, Mani-Varnosfaderani A, Benvidi A (2017) Classification of sphingosine kinase inhibitors using counter propagation artificial neural networks: a systematic route for designing selective SphK inhibitors. SAR QSAR Environ Res 28:91–109. https://doi.org/10.1080/1062936X.2017.1280535
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Chem Inf 3:1–14. https://doi.org/10.1186/1758-2946-3-33
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519. https://doi.org/10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P
Todeschini R, Consonni V, Mauri A, Pavan M (2006) DRAGONs software for the calculation of molecular descriptors, version 5.4 for Windows. Milan, Italy. http://www.talete.mi.it/products/dragon_description.htm
Boughorbel S, Jarray F, El-Anbari M (2017) Optimal classifier for imbalanced data using matthews correlation coefficient metric. PLoS ONE 12:1–17. https://doi.org/10.1371/journal.pone.0177678
Chen J, Zhou H, Aguilar A, Liu L, Bai L, McEachern D, Yang CY, Meagher L, Stuckey JA, Wang S (2012) Structure-based discovery of BM-957 as a potent small-molecule inhibitor of Bcl-2 and Bcl-xL capable of achieving complete tumor regression. J Med Chem 55(19):8502–8514. https://doi.org/10.1021/jm3010306
Zhou H, Aguilar A, Chen J, Bai L, Liu L, Meagher JL, Yang CY, McEachern D, Cong X, Stuckey JA, Wang S (2012) Structure-based design of potent Bcl-2/Bcl-xL inhibitors with strong in vivo antitumor activity. J Med Chem 55(13):6149–6161. https://doi.org/10.1021/jm300608w
Varnes JG, Gero T, Huang S, Diebold RB, Ogoe C, Grover PT, Su M, Mukherjee P, Saeh JC, MacIntyre T, Repik G (2014) Towards the next generation of dual Bcl-2/Bcl-xL inhibitors. Bioorg Med Chem Lett 24(14):3026–3033. https://doi.org/10.1016/j.bmcl.2014.05.036
Liu X, Zhang Y, Huang W, Tan W, Zhang A (2018) Design, synthesis and pharmacological evaluation of new acyl sulfonamides as potent and selective Bcl-2 inhibitors. Bioorg Med Chem 26(2):443–454. https://doi.org/10.1016/j.bmc.2017.12.001
Pellecchia M, Reed JC (2004) Inhibition of anti-apoptotic Bcl-2 family proteins by natural polyphenols new avenues for cancer chemoprevention and chemotherapy. Curr Pharm Des 10(12):1387–1398. https://doi.org/10.2174/1381612043384880
Hennessy EJ (2016) Selective inhibitors of Bcl-2 and Bcl-xL: balancing antitumor activity with on-target toxicity. Bioorg Med Chem Lett 26(9):2105–2114. https://doi.org/10.1016/j.bmcl.2016.03.032
Touré BB, Miller-Moslin K, Yusuff N, Perez L, Doré M, Joud C, Michael W, DiPietro L, van der Plas S, McEwan M, Lenoir F (2013) The role of the acidity of N-heteroaryl sulfonamides as inhibitors of Bcl-2 family protein–protein interactions. ACS Med Chem Lett 4(2):186–190. https://doi.org/10.1021/ml300321d
Rizzi A, Fioni A (2008) Virtual screening using PLS discriminant analysis and ROC curve approach: an application study on PDE4 inhibitors. J Chem Inf Model 48:1686–1692. https://doi.org/10.1021/ci800072r
Ahmed I, Greenwood R, Costello B, Ratcliffe N, Probert CS (2016) Investigation of faecal volatile organic metabolites as novel diagnostic biomarkers in inflammatory bowel disease. Aliment Pharm Therap 43(5):596–611. https://doi.org/10.1111/apt.13522
Armutlu P, Ozdemir ME, Uney-Yuksektepe F, Kavakli IH, Turkay M (2008) Classification of drug molecules considering their IC50 values using mixed-integer linear programming based hyper-boxes method. BMC Bioinform 9(1):1–14. https://doi.org/10.1186/1471-2105-9-411
Reis A, Rudnitskaya A, Chariyavilaskul P, Dhaun N, Melville V, Goddard J, Webb DJ, Pitt AR, Spickett CM (2015) Top-down lipidomics of low density lipoprotein reveal altered lipid profiles in advanced chronic kidney disease. J Lipid Res 56:413–422. https://doi.org/10.1194/jlr.M055624
Penn BS (2005) Using self-organizing maps to visualize high-dimensional data. Comput Geosci 31:531–544. https://doi.org/10.1016/j.cageo.2004.10.009
Cordel MO, Azcarraga AP (2015) Fast emulation of self-organizing maps for large datasets. Procedia Comput Sci 52:381–388. https://doi.org/10.1016/j.procs.2015.05.002
Wongravee K, Lloyd GR, Silwood CJ, Grootveld M, Brereton RG (2010) Supervised self-organizing maps for classification and determination of potentially discriminatory variables: illustrated by application to nuclear magnetic resonance metabolomic profiling. Anal Chem 82:628–638. https://doi.org/10.1021/ac9020566
Jaworska J, Nikolova-Jeliazkova N, Aldenberg T (2005) QSAR applicability domain estimation by projection of the training set descriptor space: a review. Atla-Nottingham 33(5):445 PMID:16268757
Gramatica P (2007) Principles of QSAR models validation: internal and external. Mol Inf 26(5):694–701. https://doi.org/10.1002/qsar.200610151
Randic M, Kleiner AF, De Alba LM (1994) Distance/distance matrixes. J Chem Inf Comput Sci 34:277–286. https://doi.org/10.1021/ci00018a008
Todeschini R, Consoni V (2008) Handbook of molecular descriptors. Methods and principles in medicinal chemistry. Wiley, New York. https://doi.org/10.1002/9783527613106
Plavšić D, Nikolić S, Trinajstić N, Mihalić Z (1993) On the Harary index for the characterization of chemical graphs. J Math Chem 12(1):235–250. https://doi.org/10.1007/BF01164638
Arteca GA, Lipkowitz KB, Boyd DB (2007) Molecular shape descriptors. Rev Comput Chem 9:191–253. https://doi.org/10.1002/9780470125861.ch5
Consonni V, Todeschini R, Pavan M, Gramatica P (2002) Structure/response correlations and similarity/diversity analysis by GETAWAY descriptors. 2. Application of the novel 3D molecular descriptors to QSAR/QSPR studies. J Chem Inf Comput Sci 42(3):693–705
Kier LB, Hall LH, Frazer JW (1991) An index of electrotopological state for atoms in molecules. J Math Chem 7:229–241. https://doi.org/10.1007/BF01200825
Sharma V, Goswami R, Madan AK (1997) Eccentric connectivity index: a novel highly discriminating topological descriptor for structure-property and structure-activity studies. J Chem Inf Comput Sci 37(2):273–282. https://doi.org/10.1021/ci960049h
Bounova G, de Weck O (2012) Overview of metrics and their correlation patterns for multiple-metric topology analysis on heterogeneous graph ensembles. Phys Rev E 85(1):016117–016211. https://doi.org/10.1103/PhysRevE.85.016117
Acknowledgements
Tarbiat Modares and Yazd Universities and School of Biological Sciences in IPM are greatly acknowledged for supporting this project with Grant Number BS-1395-01-11.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mani-Varnosfaderani, A., Neiband, M.S. & Benvidi, A. Identification of molecular features necessary for selective inhibition of B cell lymphoma proteins using machine learning techniques. Mol Divers 23, 55–73 (2019). https://doi.org/10.1007/s11030-018-9856-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-018-9856-x