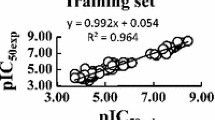
Abstract
A quantitative structure–activity (QSAR) model has been developed for enriched tubulin inhibitors, which were retrieved from sequence similarity searches and applicability domain analysis. Using partial least square (PLS) method and leave-one-out (LOO) validation approach, the model was generated with the correlation statistics of \(q^{2}\) and \(r_\mathrm{pred}^2 \) of 0.68 and 0.69, respectively. The present study indicates that topological descriptors, viz. BIC, CH_3_C, IC, JX and Kappa_2 correlate well with biological activity. ADME and toxicity (or ADME/T) assessment showed that out of 260 molecules, 255 molecules successfully passed the ADME/T assessment test, wherein the drug-likeness attributes were exhibited. These results showed that topological indices and the colchicine binding domain directly influence the aetiology of helminthic infections. Further, we anticipate that our model can be applied for guiding and designing potential anthelmintic inhibitors.





Similar content being viewed by others
Abbreviations
- MAE:
-
Mean analysis error
- PSA:
-
Polar surface area
- BBB:
-
Blood brain barrier
- WHO:
-
World Health Organization
- LDA:
-
Linear discriminant analysis
- MLR:
-
Multi linear regression
- ANN:
-
Artificial neural network
- OECD:
-
Organization for Economic Co-operation and Development
- NTDs:
-
Neglected tropical diseases
- STH:
-
Soil transmitted helminths
- nNE:
-
Number of negative prediction error value
- nPE:
-
Number of positive prediction error value
- FCFP:
-
Functional class fingerprints
- DS:
-
Discovery Studio
- SmTGR:
-
Schistosoma mansoni thioredoxin-glutathione reductase
- S-PLS:
-
Stepwise-partial least square
References
Martin RJ, Robertson AP, Bjorn H (1997) Target sites of anthelmintics. Parasitology 114:111–124
Martin RJ (1997) Modes of action of anthelmintic drugs. Vet J 154:11–34. https://doi.org/10.1016/S1090-0233(05)80005-X
Nathan ST, Mathew N, Kalyanasundaram M, Balaraman K (2005) Structure of glutathione S-transferase of the filarial parasite Wuchereria bancrofti: a target for drug development against adult worm. J Mol Model 11:194–199. https://doi.org/10.1007/s00894-005-0234-0
Plotkin S, Diemert DJ, Bethony JM, Hotez PJ (2008) Hookworm vaccines. Clin Infect Dis 46:282–288. https://doi.org/10.1016/j.vaccine.2012.11.034
World Health Organization (2009) Neglected tropical diseases, hidden successes, emerging opportunities. http://apps.who.int/iris/bitstream/10665/44214/1/9789241598705_eng.pdf. Accessed date 2/06/2017
Relf VE, Lester HE, Morgan ER, Hodgkinson JE, Matthews JB (2014) Anthelmintic efficacy on UK Thoroughbred stud farms. Int J Parasitol 44:507–514. https://doi.org/10.1016/j.ijpara.2014.03.006
Ranjan P, Kumar SP, Kari V, Jha PC (2017) Exploration of interaction zones of \(\beta \)-tubulin colchicine binding domain of helminths and binding mechanism of anthelmintics. Comput Biol Chem 68:78–91. https://doi.org/10.1016/j.compbiolchem.2017.02.008
Leach AR, Shoichet BK, Peishoff CE (2006) Prediction of protein–ligand interactions. Docking and scoring: successes and gaps. J Med Chem 49:5851–5855. https://doi.org/10.1021/jm060999m
Charan KP, Ranjan P, Manojkumar K, Pothanagandhi N, Jha PC, Khedkar VM, Sivaramakrishna A, Vijayakrishna K (2015) Evaluation of imidazolium-based ionic liquids towards vermicidal activity: in vitro & in silico studies. RSC Adv 5:75415–75424. https://doi.org/10.1039/C5RA13469B
Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK (2002) Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem 45:2213–2221. https://doi.org/10.1021/jm010548w
Montero-Torres A, Vega MC, Marrero-Ponce Y, Rolón M, Gómez-Barrio A, Escario JA, Arán VJ, Martínez-Fernández AR, Meneses-Marcel A (2005) A novel non-stochastic quadratic fingerprints-based approach for the ‘in silico’ discovery of new antitrypanosomal compounds. Bioorgan Med Chem 13:6264–6275. https://doi.org/10.1016/j.bmc.2005.06.049
Martin MB, Sanders JM, Kendrick H, de Luca-Fradley K, Lewis JC, Grimley JS, Van Brussel EM, Olsen JR, Meints GA, Burzynska A (2002) Activity of bisphosphonates against Trypanosoma brucei rhodesiense. J Med Chem 45:2904–2914. https://doi.org/10.1021/jm0102809
Prado-Prado FJ, González-Díaz H, de la Vega OM, Ubeira FM, Chou K-C (2008) Unified QSAR approach to antimicrobials. Part 3: first multi-tasking QSAR model for input-coded prediction, structural back-projection, and complex networks clustering of antiprotozoal compounds. Bioorgan Med Chem 16:5871–5880. https://doi.org/10.1016/j.bmc.2008.04.068
Prado-Prado FJ, García-Mera X, González-Díaz H (2010) Multi-target spectral moment QSAR versus ANN for antiparasitic drugs against different parasite species. Bioorgan Med Chem 18:2225–2231. https://doi.org/10.1016/j.bmc.2010.01.068
Chou K-C, Wei D-Q, Du Q-S, Sirois S, Zhong W-Z (2006) Progress in computational approach to drug development against SARS. Curr Med Chem 13:3263–3270. https://doi.org/10.2174/092986706778773077
Chou K-C (2004) Structural bioinformatics and its impact to biomedical science. Curr Med Chem 11:2105–2134. https://doi.org/10.2174/0929867043364667
Grassy G, Calas B, Yasri A, Lahana R, Woo J, Iyer S, Kaczorek M, Floc’h R, Buelow R (1998) Computer-assisted rational design of immunosuppressive compounds. Nat Biotechnol 16:748–752. https://doi.org/10.1038/nbt0898-748
Athar M, Lone MY, Khedkar VM, Jha PC (2016) Pharmacophore model prediction, 3D-QSAR and molecular docking studies on vinyl sulfones targeting Nrf2-mediated gene transcription intended for anti-Parkinson drug design. J Biomol Struct Dyn 34:1282–1297. https://doi.org/10.1080/07391102.2015.1077343
Dearden JC (2016) The history and development of quantitative structure–activity relationships (QSARs). Oncology: breakthroughs in research and practice: breakthroughs in research and practice, p 67. https://doi.org/10.1517/17460441.2015.1083006
Rekker RF (1977) The hydrophobic fragmental constant, its derivation and application: a means of characterizing membrane systems, Pharmacochemistry library, vol 1. Elsevier, New York. ISBN 0444415483 9780444415486
Athar M, Lone MY, Jha PC (2017) First protein drug target’s appraisal of lead-likeness descriptors to unfold the intervening chemical space. J Mol Graph Model 72:272–282. https://doi.org/10.1016/j.jmgm.2016.12.019
Hansch C, Leo A, Hoekman D (1995) Exploring QSAR, ACS Professional Reference Book. ACS, Washington, DC. https://doi.org/10.1021/jm950902o
Dube D, Periwal V, Kumar M, Sharma S, Singh TP, Kaur P (2012) 3D-QSAR based pharmacophore modeling and virtual screening for identification of novel pteridine reductase inhibitors. J Mol Model 18:1701–1711. https://doi.org/10.1007/s00894-011-1187-0
Yasri A, Hartsough D (2001) Toward an optimal procedure for variable selection and QSAR model building. J Chem Inf Model 41:1218–1227. https://doi.org/10.1021/ci010291a
Todeschini R, Consonni V (2009) Molecular descriptors for chemoinformatics: volume I: alphabetical listing/volume II: appendices, references, vol 41. Wiley, Weinheim. https://doi.org/10.1002/9783527628766
Kier LB, Hall LH (1986) Molecular connectivity in structure–activity analysis. Research Studies Press-Wiley, Chichester. https://doi.org/10.1016/0160-9327(86)90116-X
Tomczak J (2003) Data types. In: Gasteiger J (ed) Handbook of chemoinformatics: from data to knowledge in 4 volumes. Wiley, Weinheim. https://doi.org/10.1002/9783527618279.ch13
Mattioni BE, Jurs PC (2002) Development of quantitative structure–activity relationship and classification models for a set of carbonic anhydrase inhibitors. J Chem Inf Model 42:94–102. https://doi.org/10.1021/ci0100696
Yang Y-Q, Xu L, Hu C-Y (1994) Extended adjacency matrix indices and their applications. J Chem Inf Model 34:1140–1145. https://doi.org/10.1021/ci00021a020
Balaban AT (1995) Chemical graphs: looking back and glimpsing ahead. J Chem Inf Model 35:339–350. https://doi.org/10.1021/ci00025a001
Ivanciuc O, Ivanciuc T, Klein D, Seitz W, Balaban A (2001) Quantitative structure-retention relationships for gas chromatographic retention indices of alkylbenzenes with molecular graph descriptors. SAR QSAR Environ Res 11:419–452. https://doi.org/10.1080/10629360108035362
Estrada E (1999) Connectivity polynomial and long-range contributions in the molecular connectivity model. Chem Phys Lett 312:556–560. https://doi.org/10.1016/S0009-2614(99)01007-6
Zhang S-X, Feng J, Kuo S-C, Brossi A, Hamel E, Tropsha A, Lee K-H (2000) Antitumor Agents. 199.Three-dimensional quantitative structure–activity relationship study of the colchicine binding site ligands using comparative molecular field analysis. J Med Chem 43:167–176. https://doi.org/10.1021/jm990333a
Fortin S, Labrie P, Moreau E, Wei L, Kotra LP, René C (2008) A comparative molecular field and comparative molecular similarity indices analyses (CoMFA and CoMSIA) of N-phenyl-N\(^\prime \)-(2-chloroethyl) ureas targeting the colchicine-binding site as anticancer agents. Bioorgan Med Chem 16:1914–1926. https://doi.org/10.1016/j.bmc.2007.11.004
Polański J (2000) The non-grid technique for modeling 3D QSAR using self-organizing neural network (SOM) and PLS analysis: application to steroids and colchicinoids. SAR QSAR Environ Res 11:245–261. https://doi.org/10.1080/10629360008033234
Sanders JM, Gómez AO, Mao J, Meints GA, Van Brussel EM, Burzynska A, Kafarski P, González-Pacanowska D, Oldfield E (2003) 3-D QSAR investigations of the inhibition of Leishmania major farnesyl pyrophosphate synthase by bisphosphonates. J Med Chem 46:5171–5183. https://doi.org/10.1021/jm0302344
Ducki S, Mackenzie G, Lawrence NJ, Snyder JP (2005) Quantitative structure–activity relationship (5D-QSAR) study of combretastatin-like analogues as inhibitors of tubulin assembly. J Med Chem 48:457–465. https://doi.org/10.1021/jm049444m
Kumar SP, Jha PC, Jasrai YT, Pandya HA (2016) The effect of various atomic partial charge schemes to elucidate consensus activity-correlating molecular regions: a test case of diverse QSAR models. J Biomol Struct Dyn 34:540–559. https://doi.org/10.1080/07391102.2015.1044474
Vega MC, Montero-Torres A, Marrero-Ponce Y, Rolón M, Gómez-Barrio A, Escario JA, Arán VJ, Nogal JJ, Meneses-Marcel A, Torrens F (2006) New ligand-based approach for the discovery of antitrypanosomal compounds. Bioorgan Med Lett 16:1898–1904. https://doi.org/10.1016/j.bmcl.2005.12.087
Lima CR, Carels N, Guimaraes ACR, Tufféry P, Derreumaux P (2016) In silico structural characterization of protein targets for drug development against Trypanosoma cruzi. J Mol Model 22:244. https://doi.org/10.1007/s00894-016-3115-9
Castillo-Garit JA, Vega MC, Rolón M, Marrero-Ponce Y, Gómez-Barrio A, Escario JA, Bello AA, Montero A, Torrens F, Pérez-Giménez F (2011) Ligand-based discovery of novel trypanosomicidal drug-like compounds: in silico identification and experimental support. Eur J Med Chem 46:3324–3330. https://doi.org/10.1016/j.ejmech.2011.04.057
Montresor A, Gabrielli AF, Yajima A, Lethanh N, Biggs B-A, Casey GJ, Tinh TT, Engels D, Savioli L (2013) Markov model to forecast the change in prevalence of soil-transmitted helminths during a control programme: a case study in Vietnam. Trans R Soc Trop Med Hyg 107:313–318. https://doi.org/10.1093/trstmh/trt019
Lipkowitz K, McCracken R (1993) Molecular modeling: a tool for predicting anthelmintic activity in vivo. Parasitol Res 79:475–479. https://doi.org/10.1007/BF00931586
Melo-Filho CC, Dantas RF, Braga RC, Neves BJ, Senger MR, Valente WC, Rezende-Neto JM, Chaves WT, Muratov EN, Paveley RA (2016) Qsar-driven discovery of novel chemical scaffolds active against Schistosoma mansoni. J Chem Inf Model 56:1357–1372. https://doi.org/10.1021/acs.jcim.6b00055
Castillo-Garit JA, del Toro-Cortés O, Kouznetsov VV, Puentes CO, Romero Bohórquez AR, Vega MC, Rolón M, Escario JA, Gómez-Barrio A, Marrero-Ponce Y (2012) Identification in-silico and in vitro of novel trypanosomicidal drug-like compounds. Chem Biol Drug Des 80:38–45. https://doi.org/10.1111/j.1747-0285.2012.01378.x
Marrero-Ponce Y, Romero V (2002) TOMOCOMD software. Universidad Tecnológica de Bolívar, Colombia
Mohamadi F, Richards NG, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC (1990) MacroModel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem 11:440–467. https://doi.org/10.1002/jcc.540110405
Marrero-Ponce Y, Castillo-Garit JA, Olazabal E, Serrano HS, Morales A, Castanedo N, Ibarra-Velarde F, Huesca-Guillen A, Sánchez AM, Torrens F (2005) Atom, atom-type and total molecular linear indices as a promising approach for bioorganic and medicinal chemistry: theoretical and experimental assessment of a novel method for virtual screening and rational design of new lead anthelmintic. Bioorgan Med Chem 13:1005–1020. https://doi.org/10.1016/j.bmc.2004.11.040
Ponce YM (2003) Total and local quadratic indices of the molecular pseudograph’s atom adjacency matrix: applications to the prediction of physical properties of organic compounds. Molecules 8:687–726. https://doi.org/10.3390/80900687
Ponce YM, Perez MC, Zaldivar VR, Diaz HG, Torrens F (2004) A new topological descriptors based model for predicting intestinal epithelial transport of drugs in Caco-2 cell culture. J Pharm Pharm Sci 7:186–199
Escher BI, Berger C, Bramaz N, Kwon JH, Richter M, Tsinman O, Avdeef A (2008) Membrane–water partitioning, membrane permeability, and baseline toxicity of the parasiticides ivermectin, albendazole, and morantel. Environ Toxicol Chem 27:909–918. https://doi.org/10.1897/07-427.1
Chan C, Yin H, Garforth J, McKie JH, Jaouhari R, Speers P, Douglas KT, Rock PJ, Yardley V, Croft SL (1998) Phenothiazine inhibitors of trypanothione reductase as potential antitrypanosomal and antileishmanial drugs. J Med Chem 41:148–156. https://doi.org/10.1021/jm960814j
Borrego-Sánchez A, Hernández-Laguna A, Sainz-Díaz CI (2017) Molecular modeling and infrared and Raman spectroscopy of the crystal structure of the chiral antiparasitic drug Praziquantel. J Mol Model 23:106. https://doi.org/10.1007/s00894-017-3266-3
Sanches SM, Taft CA (2004) A molecular modeling and QSAR study of suppressors of the growth of Trypanosoma cruzi epimastigotes. J Mol Graph Model 23:89–97. https://doi.org/10.1016/j.jmgm.2004.03.013
Prieto JJ, Talevi A, Bruno-Blanch LE (2006) Application of linear discriminant analysis in the virtual screening of antichagasic drugs through trypanothione reductase inhibition. Mol Divers 10:361–375. https://doi.org/10.1007/s11030-006-9044-2
Planche AS, Scotti MT, Emerenciano VDP, López AG, Pérez EM, Uriarte E (2010) Designing novel antitrypanosomal agents from a mixed graph-theoretical substructural approach. J Comput Chem 31:882–894. https://doi.org/10.1002/jcc.21374
Neves BJ, Braga RC, Bezerra JC, Cravo PV, Andrade CH (2015) In silico repositioning-chemogenomics strategy identifies new drugs with potential activity against multiple life stages of Schistosoma mansoni. PLoS Negl Trop Dis 9:e3435. https://doi.org/10.1371/journal.pntd.0003435
Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, Holroyd N, Bartley DJ, Beasley H, Britton C (2013) The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol 14:R88. https://doi.org/10.1186/gb-2013-14-8-r88
Redman E, Sargison N, Whitelaw F, Jackson F, Morrison A, Bartley DJ, Gilleard JS (2012) Introgression of ivermectin resistance genes into a susceptible Haemonchus contortus strain by multiple backcrossing. PLoS Pathog 8:e1002534. https://doi.org/10.1371/journal.ppat.1002534
Holden-Dye L, Walker R (2014) Anthelmintic drugs and nematocides: studies in Caenorhabditis elegans. WormBook: the online review of C. elegans biology, 1–29. https://doi.org/10.1895/wormbook.1.143.2
Schwarz EM, Korhonen PK, Campbell BE, Young ND, Jex AR, Jabbar A, Hall RS, Mondal A, Howe AC, Pell J (2013) The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol 14:R89. https://doi.org/10.1186/gb-2013-14-8-r89
Laing R, Hunt M, Protasio AV, Saunders G, Mungall K, Laing S, Jackson F, Quail M, Beech R, Berriman M (2011) Annotation of two large contiguous regions from the Haemonchus contortus genome using RNA-seq and comparative analysis with Caenorhabditis elegans. PLoS One 6:e23216. https://doi.org/10.1371/journal.pone.0023216
Gilleard JS (2013) Haemonchus contortus as a paradigm and model to study anthelmintic drug resistance. Parasitolgy 140:1506–1522. https://doi.org/10.1017/S0031182013001145
Gilleard JS (2006) Understanding anthelmintic resistance: the need for genomics and genetics. Int J Parasitol 36:1227–1239. https://doi.org/10.1016/j.ijpara.2006.06.010
Kaminsky R, Gauvry N, Weber SS, Skripsky T, Bouvier J, Wenger A, Schroeder F, Desaules Y, Hotz R, Goebel T (2008) Identification of the amino-acetonitrile derivative monepantel (AAD 1566) as a new anthelmintic drug development candidate. Parasitol Res 103:931–939. https://doi.org/10.1007/s00436-008-1080-7
LeJambre LF, Windon RG, Smith WD (2008) Vaccination against Haemonchus contortus: performance of native parasite gut membrane glycoproteins in Merino lambs grazing contaminated pasture. Vet Parasitol 153:302–312. https://doi.org/10.1016/j.vetpar.2008.01.032
Taylor CM, Martin J, Rao RU, Powell K, Abubucker S, Mitreva M (2013) Using existing drugs as leads for broad spectrum anthelmintics targeting protein kinases. PLoS Pathog 9:e1003149. https://doi.org/10.1371/journal.ppat.1003149
Meier L, Torgerson PR, Hertzberg H (2016) Vaccination of goats against Haemonchus contortus with the gut membrane proteins H11/H-gal-GP. Vet Parasitol 229:15–21. https://doi.org/10.1016/j.vetpar.2016.08.024
Bethony J, Loukas A, Hotez P, Knox D (2006) Vaccines against blood-feeding nematodes of humans and livestock. Parasitology 133:S63–S79. https://doi.org/10.1017/S0031182006001818
Robinson M, Trudgett A, Fairweather I, McFerran N (2002) Benzimidazole binding to Haemonchus contortus tubulin: a question of structure. Trends Parasitol 18:153–154. https://doi.org/10.1016/S1471-4922(01)02225-5
Robinson MW, McFerran N, Trudgett A, Hoey L, Fairweather I (2004) A possible model of benzimidazole binding to \(\beta \)-tubulin disclosed by invoking an inter-domain movement. J Mol Graph Model 23:275–284. https://doi.org/10.1016/j.jmgm.2004.08.001
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank, 1999-. In: Rossmann MG, Arnold E (eds) International tables for crystallography volume F: crystallography of biological macromolecules. Springer, Netherlands, Amsterdam
Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. https://doi.org/10.1093/nar/gkq366
Holm L, Kääriäinen S, Rosenström P, Schenkel A (2008) Searching protein structure databases with DaliLite v. 3. Bioinformatics 24:2780–2781. https://doi.org/10.1093/bioinformatics/btn507
Holm L, Park J (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16:566–567. https://doi.org/10.1093/bioinformatics/16.6.566
Holm L, Sander C (1994) Searching protein structure databases has come of age. Proteins 19:165–173. https://doi.org/10.1002/prot.340190302
Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic acids Res 3:W590–W594. https://doi.org/10.1093/nar/gkh477
Studio Discovery, version 4.0, (2013) Accelrys. San Diego, USA
Roy K, Kar S, Das RN (2015) A primer on QSAR/QSPR modeling: fundamental concepts. Springer, New York. https://doi.org/10.1007/978-3-319-17281-1_1
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217. https://doi.org/10.1002/jcc.540040211
Karelson M (2000) Molecular descriptors in QSAR/QSPR. Wiley, New York. ISBN 978-0-471-35168-9
Gobbi A, Lee M-L (2003) DISE: directed sphere exclusion. J Chem Inf Model 43:317–323. https://doi.org/10.1021/ci025554v
Roy K, Kar S, Ambure P (2015) On a simple approach for determining applicability domain of QSAR models. Chemom Intell Lab 145:22–29. https://doi.org/10.1016/j.chemolab.2015.04.013
Krishnaiah PR (2014) Multivariate Analysis—III: Proceedings of the Third International Symposium on Multivariate Analysis Held at Wright State University, Dayton, Ohio, June 19–24, 1972. Academic Press
Wold S (1995) Chemometrics; what do we mean with it, and what do we want from it? Chemom Intell Lab 30:109–115 ISBN 1483265137, 9781483265131
Kulkarni SS, Kulkarni VM (1999) Three-dimensional quantitative structure–activity relationship of interleukin 1-\(\beta \) converting enzyme inhibitors: a comparative molecular field analysis study. J Med Chem 42:373–380. https://doi.org/10.1021/jm9708442
Refaeilzadeh P, Tang L, Liu H (2009) Cross-validation. In: Liu L, Tamer Özsu M (eds) Encyclopedia of database systems. Springer, New York, pp 532–538. https://doi.org/10.1007/978-0-387-39940-9_56
Stone M (1974) Cross-validatory choice and assessment of statistical predictions. J R Stat Soc B Met B 36:111–147
Roy K (2016) Chemoinformatics tools. http://dtclab.webs.com/software-tools. Accessed 20 May 2017
Roy K, Mitra I, Kar S, Ojha PK, Das RN, Kabir H (2012) Comparative studies on some metrics for external validation of QSPR models. J Chem Inf Model 52:396–408. https://doi.org/10.1021/ci200520g
Roy K, Ambure P (2016) The “double cross-validation” software tool for MLR QSAR model development. Chemom Intell Lab 159:108–126. https://doi.org/10.1016/j.chemolab.2016.10.009
Sharma MC, Sharma S, Sahu NK, Kohli D (2013) QSAR studies of some substituted imidazolinones angiotensin II receptor antagonists using partial least squares regression (PLSR) method based feature selection. J Saudi Chem Soc 17:219–225. https://doi.org/10.1016/j.jscs.2011.03.012
Cheng A, Merz KM (2003) Prediction of aqueous solubility of a diverse set of compounds using quantitative structure–property relationships. J Med Chem 46:3572–3580. https://doi.org/10.1021/jm020266b
Egan WJ, Lauri G (2002) Prediction of intestinal permeability. Adv Drug Deliv Rev 54:273–289. https://doi.org/10.1016/S0169-409X(02)00004-2
Egan WJ, Merz KM, Baldwin JJ (2000) Prediction of drug absorption using multivariate statistics. J Med Chem 43:3867–3877. https://doi.org/10.1021/jm000292e
Susnow RG, Dixon SL (2003) Use of robust classification techniques for the prediction of human cytochrome P450 2D6 inhibition. J Chem Inf Model 43:1308–1315. https://doi.org/10.1021/ci030283p
Dixon SL, Merz KM (2001) One-dimensional molecular representations and similarity calculations: methodology and validation. J Med Chem 44:3795–3809. https://doi.org/10.1021/jm010137f
Votano JR, Parham M, Hall LM, Hall LH, Kier LB, Oloff S, Tropsha A (2006) QSAR modeling of human serum protein binding with several modeling techniques utilizing structure–information representation. J Med Chem 49:7169–7181. https://doi.org/10.1021/jm051245v
Xia X, Maliski EG, Gallant P, Rogers D (2004) Classification of kinase inhibitors using a Bayesian model. J Med Chem 47:4463–4470. https://doi.org/10.1021/jm0303195
Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M (2004) Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428:198–202. https://doi.org/10.1038/nature02393
Dorléans A, Gigant B, Ravelli RB, Mailliet P, Mikol V, Knossow M (2009) Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. P Natl Acad Sci Biol 106:13775–13779. https://doi.org/10.1073/pnas.0904223106
Abdi H (2010) Partial least squares regression and projection on latent structure regression (PLS Regression). Wiley Interdiscip Rev Comput Stat 2:97–106. https://doi.org/10.1002/wics.051
Roy K, Kar S, Das RN (2015) Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment. Academic Press, San Diego eBook
Alberto Castillo-Garit J, Abad C, Enrique Rodriguez-Borges J, Marrero-Ponce Y, Torrens F (2012) A review of QSAR studies to discover new drug-like compounds actives against leishmaniasis and trypanosomiasis. Curr Top Med Chem 12:852–865. https://doi.org/10.2174/156802612800166756
Roy K, Das RN, Ambure P, Aher RB (2016) Be aware of error measures. Further studies on validation of predictive QSAR models. Chemom Intell Lab 152:18–33. https://doi.org/10.1016/j.chemolab.2016.01.008
Roy K, Chakraborty P, Mitra I, Ojha PK, Kar S, Das RN (2013) Some case studies on application of "\(r_m^2\)" metrics for judging quality of quantitative structure–activity relationship predictions: emphasis on scaling of response data. J Comput Chem 34:1071–1082. https://doi.org/10.1002/jcc.23231
Consonni V, Ballabio D, Todeschini R (2010) Evaluation of model predictive ability by external validation techniques. J Chemom 24:194–201. https://doi.org/10.1002/cem.1290
Hou T, Xu X (2002) ADME evaluation in drug discovery. J Mol Model 8:337–349. https://doi.org/10.1007/s00894-002-0101-1
Remya C, Dileep KV, Tintu I, Variyar EJ, Sadasivan C (2013) In vitro inhibitory profile of NDGA against AChE and its in silico structural modifications based on ADME profile. J Mol Model 19:1179–1194. https://doi.org/10.1007/s00894-002-0101-1
Acknowledgements
Prabodh Ranjan would like to thank the financial support from the University Grants Commission (UGC), Govt. of India. Mohd Athar acknowledges generous support from the Department of Science and Technology (DST), Govt. of India as INSPIRE-SRF Fellowship. Prakash C. Jha also would like to thank UGC for providing SERB (EMR/2016/003025) Grant, and the Central University of Gujarat for providing basic computational facilities. Discussion with Dr. Pravin Ambure, University of Gdansk, Poland are also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors have no personal, financial or non-financial conflicts of interest regarding the publication of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ranjan, P., Athar, M., Jha, P.C. et al. Probing the opportunities for designing anthelmintic leads by sub-structural topology-based QSAR modelling. Mol Divers 22, 669–683 (2018). https://doi.org/10.1007/s11030-018-9825-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-018-9825-4