Abstract
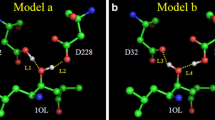
Mercury(II) has a strong affinity for the thiol groups in proteins often resulting in the disruption of their biological functions. In this study we present classical and first-principles, DFT-based molecular dynamics (MD) simulations of a complex of Hg(II) and proteinase K, a well-known serine protease with a very broad and diverse enzymatic activity. It contains a catalytic triad formed by Asp39, His69, and Ser224, which is responsible for its biological activity. It was found previously by X-ray diffraction experiments that the presence of Hg(II) inhibits the enzymatic action of proteinase K by affecting the stereochemistry of the triad. Our simulations predict that (i) the overall structure as well as the protein backbone dynamics are only slightly affected by the mercury cation, (ii) depending on the occupied mercury site, the hydrogen bonds of the catalytic triad are either severely disrupted (both bonds for mercury at site 1, and the His69–Ser224 contact for mercury at site 2) or slightly strengthened (the Asp39–His69 bond when mercury is at site 2), (iii) the network of hydrogen bonds of the catalytic triad is not static but undergoes constant fluctuations, which are significantly modified by the presence of the Hg(II) cation, influencing in turn the triad’s ability to carry out the enzymatic function—these facts explain the experimental findings on the inhibition of proteinase K by Hg(II).
Similar content being viewed by others
Abbreviations
- BOMD:
-
Born-Oppenheimer molecular dynamics
- DFT:
-
Density functional theory
- MD:
-
Molecular dynamics
- QTAIM:
-
Quantum Theory of Atoms in Molecules
- PDB:
-
Protein Data Bank
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- RGYR:
-
Radius of gyration
- SASA:
-
Solvent-accessible surface area
References
D’Itri P (1977) Mercury contamination: a human tragedy. Wiley, New York
Jiang G-B, Shi J-B, Feng X-B (2006) Mercury pollution in China. Environ Sci Technol 40: 3672–3678. doi:10.1021/es062707c
Devlin EW (2006) Acute toxicity, uptake and histopathology of aqueous methyl mercury to fathead minnow embryos. Ecotoxicology 15: 97–110. doi:10.1007/s10646-005-0051-3
Onyido I, Norris AR, Buncel E (2004) Biomolecule–mercury interactions: modalities of DNA base-mercury binding mechanisms. Remediation strategies. Chem Rev 104: 5911–5929. doi:10.1021/cr030443w
Okino S, Iwasaki K, Yagi O, Tanaka H (2000) Development of a biological mercury removal-recovery system. Biotechnol Lett 22: 783–788
Sigel A, Sigel H (1997) Metal ions in biological systems. Marcel Dekker, New York
Spiro TG (1980) Nucleic acid–metal ion interactions. Wiley, New York
Eichhorn GL (1973) Inorganic biochemistry, vol 2, chap 33. Elsevier, Amsterdam, p 1210
Kurzel RB, Cetrulo CL (1981) Critical review. The effect of environmental pollutants on human reproduction, including birth defects. Environ Sci Technol 15: 626–640. doi:10.1021/es00088a001
Zahir F, Rizwi SJ, Haq SK, Khan RH (2005) Low dose mercury toxicity and human health. Environ Toxicol Pharmacol 20: 351–360. doi:10.1016/j.etap.2005.03.007
Saxena AK, Singh TP, Peters K, Fittkau S, Visanji M, Wilson KS, Betzel C (1996) Structure of a ternary complex of proteinase K, mercury, and a substrate-analogue hexa-peptide at 2.2 Å resolution. Proteins Struct Funct Genet 25: 195–201
Ebeling W, Hennrich N, Klockow M, Metz H, Orth HD, Lang H (1974) Proteinase K from Tritirachium album Limber. Eur J Biochem 47: 91–97
Jany K-D, Lederer G, Mayer B (1986) Amino acid sequence of proteinase K from the mold Tritirachium album Limber: proteinase K—a subtilisin-related enzyme with disulfide bonds. FEBS Lett 199: 139–144. doi:10.1016/0014-5793(86)80467-7
Müller A, Saenger W (1993) Studies on the inhibitory action of mercury upon proteinase K. J Biol Chem 268: 26150–26154
Betzel C, Gourinath S, Kumar P, Kaur P, Perbandt M, Eschenburg S, Singh TP (2001) Structure of a serine protease Proteinase K from Tritirachium album Limber at 0.98 Å resolution. Biochemistry 40: 3080–3088. doi:10.1021/bi002538n
Neurath H (1986) The versatility of proteolytic enzymes. J Cell Biochem 32: 35–49. doi:10.1002/jcb.240320105
Russel AJ, Fersht AR (1987) Rational modification of enzyme catalysis by engineering surface charge. Nature 328: 496–500. doi:10.1038/328496a0
Ni B, Kramer JR, Bell RA, Werstiuk NH (2006) Protonolysis of the Hg–C bond of chloromethylmercury and dimethylmercury. A DFT and QTAIM study. J Phys Chem A 110: 9451–9458. doi:10.1021/jp061852+
Tai H-C, Lim C (2006) Computational studies of the coordination stereochemistry, bonding, and metal selectivity of mercury. J Phys Chem A 110: 452–462. doi:10.1021/jp0529826
Marx D, Hutter J (2000) In Grotendorst J (ed) NIC Series, vol 1, Modern methods and algorithms of quantum chemistry. John von Neuman Institute for Computing, FZ Jülich, pp 329–477. http://www.fz-juelich.de/nic-series/Volume1/
VandeVondele J, Krack M, Mohamed F, Parrinello M, Chassaing T, Hutter J (2005) Quickstep: fast and accurate density functional calculations using a mixed Gaussian and plane waves approach. Comput Phys Commun 167: 103–128. doi:10.1016/j.cpc.2004.12.014
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98: 5648–5652. doi:10.1063/1.464913
Lee C, Yang W, Parr RG (1988) Development of the Colle–Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37: 785–789. doi:10.1103/PhysRevB.37.785
Andrae D, Häussermann U, Dolg M, Stoll H, Preuss H (1990) Energy-adjusted ab initio pseudopotentials for the second and third row transition elements. Theor Chim Acta 77: 123–141. doi:10.1007/BF01114537
Besler BH, Merz KMJr, Kollman PA (1990) Atomic charges derived from semiempirical methods. J Comput Chem 11: 431–439. doi:10.1002/jcc.540110404
Gaussian 03, Revision C.02, Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian, Inc., Wallingford, CT, USA
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935. doi:10.1063/1.445869
Case DA, Darden TA, Cheatham III TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006) AMBER 9, University of California, San Francisco
Darden T, York D, Pedersen LG (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys 98: 10089–10092. doi:10.1063/1.464397
Essman U, Perela L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103: 8577–8592. doi:10.1063/1.470117
Sagui C, Darden T (1999) Molecular dynamics simulations of biomolecules: long-range electrostatic effects. Annu Rev Biophys Biomol Struct 28: 155–179. doi:10.1146/annurev.biophys.28.1.155
Adelman SA, Doll JD (1976) Generalized Langevin equation approach for atom/solid-surface scattering: general formulation for classical scattering off harmonic solids. J Chem Phys 64: 2375–2388. doi:10.1063/1.432526
Feller SE, Zhang Y, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103: 4613–4621. doi:10.1063/1.470648
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23: 327–341. doi:10.1016/0021-9991(77)90098-5
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–38. doi:10.1016/0263-7855(96)00018-5
Lippert G, Hutter J, Parrinello M (1997) A hybrid Gaussian and plane wave density functional scheme. Mol Phys 92: 477–487. doi:10.1080/00268979709482119
Genovese L, Deutsch T, Neelov A, Goedecker S, Beylkin G (2006) Efficient solution of Poisson’s equation with free boundary conditions. J Chem Phys 125: 074105. doi:10.1063/1.2335442
Goedecker S, Teter M, Hutter J (1996) Separable dual-space Gaussian pseudopotentials. Phys Rev B 54: 1703–1710. doi:10.1103/PhysRevB.54.1703
Hartwigsen J, Goedecker S, Hutter J (1998) Relativistic separable dual-space Gaussian pseudopotentials from H to Rn. Phys Rev B 58: 3641–3662. doi:10.1103/PhysRevB.58.3641
Krack M (2005) Pseudopotentials for H to Kr optimized for gradient-corrected exchange-correlation functionals. Theor Chem Acc 114: 145–152. doi:10.1007/s00214-005-0655-y
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A 38: 3098–3100. doi:10.1103/PhysRevA.38.3098
Nosé S (1984) A molecular dynamics method for simulations in the canonical ensemble. Mol Phys 52: 255–268. doi:10.1080/00268978400101201
Nosé S (1984) A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys 81: 511–519. doi:10.1063/1.447334
Mulliken RS (1955) Electronic population analysis on LCAO-MO molecular wave functions. J Chem Phys 23: 1833–1840. doi:10.1063/1.1740588
CP2K version 2.0.0 (Development Version), the CP2K developers group (2008) CP2K is freely available from http://cp2k.berlios.de/
Gnuplot 4.0. Copyright © 1986–1993, 1998, 2004, Williams T, Kelley C
Herrera FE, Zucchelli S, Jezierska A, Lavina ZS, Gustincich S, Carloni P (2007) On the oligomeric state of DJ-1 protein and its mutants associated with Parkinson disease: a combined computational and in vitro study. J Biol Chem 282: 24905–24914. doi:10.1074/jbc.M701013200
Marx D (2006) Proton transfer 200 years after von Grotthuss: insights from ab initio simulations. ChemPhysChem 7: 1848–1870. doi:10.1002/cphc.200600128
Betzel C, Pal GP, Saenger W (1988) Three-dimensional structure of proteinase K at 0.15-nm resolution. Eur J Biochem 178: 155–171. doi:10.1111/j.1432-1033.1988.tb14440.x
Ertekin A, Nussinov R, Haliloglu T (2006) Association of putative concave protein-binding sites with the fluctuation behavior of residues. Protein Sci 15: 2265–2277. doi:10.1110/ps.051815006
Kalosakas G, Rasmussen KØ, Bishop AR, Choi CH, Usheva A (2004) Sequence-specific thermal fluctuations identify start sites for DNA transcription. Europhys Lett 68: 127–133. doi:10.1209/epl/i2004-10167-8
Pordea A, Creus M, Panek J, Duboc C, Mathis D, Novič M, Ward TR (2008) Artificial metalloenzyme for enantioselective sulfoxidation based on vanadyl-loaded streptavidin. J Am Chem Soc 130: 8085–8088. doi:10.1021/ja8017219
Miño G, Contreras R (2009) On the role of short and strong hydrogen bonds on the mechanism of action of a model chymotrypsine active site. J Phys Chem A 113: 5769–5772. doi:10.1021/jp902756x
Barkay T, Miller SM, Summers AO (2003) Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27: 355–384. doi:10.1016/S0168-6445(03)00046-9
Howell SC, Mesleh MF, Opella SJ (2005) NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry 44: 5196–5206. doi:10.1021/bi048095v
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Panek, J.J., Mazzarello, R., Novič, M. et al. Impact of Mercury(II) on proteinase K catalytic center: investigations via classical and Born-Oppenheimer molecular dynamics. Mol Divers 15, 215–226 (2011). https://doi.org/10.1007/s11030-010-9256-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-010-9256-3