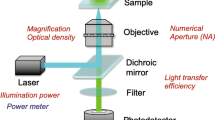
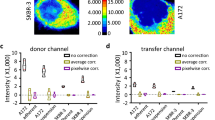
Abstract
Fluorescence image analysis provides quantitative data on fluorescence in situ hybridiza-tion signals (FISH), immunofluorescence labelings, Green Fluorescent Protein (GFP) expression and microarrays. It is a valuable tool for decision making in the fields of biology and medicine. The aim of this study was to evaluate the reproducibility of fluorescence intensity measurements and standardization when acquisitions are performed under various but well defined conditions. Fluorescent intensity of standard beads (Inspeck series, Molecular Probes) was repeatedly measured using an image analyzer and automated procedures. Images were acquired using several integration times and neutral filter sets. A standardization procedure was used for expressing the data in a same unit: data were multiplied by the light attenuation factor and were divided by the CCD integration times. Results show that 1) standardiza-tion is possible 2) accurate and reliable fluorescence measurements can be obtained and 3) specimens showing large differences in fluorescence intensity can be objectively compared. Moreover fluorescent test slides including fluorochrome solutions and altuglas slides were tested for shading correction and as overall test systems.
Similar content being viewed by others
References
Bezakova G, Helm JP, Francolini M, Lomo T (2001). Effects of purified recombinant neural and muscle agrin on skeletal muscle fibers in vivo. J Cell Biol 153: 1441–1452
Bour-Dill C, Gramain MP, Merlin JL, Marchal S, Guillemin F (2000). Determination of intracellular organelles implicated in daunorubicin cytoplasmic sequestration in multidrug-resistant MCF-7 cells using fluorescence microscopy image analysis. Cytometry 39: 16–25
Forster T, Roy D, Ghazal P (2003). Experiments using microarray technology: limitations and standard oper-ating procedures. J Endocrinol 178: 195–204
Holmes KL, Lantz LM (2001). Protein labeling with fluorescent probes. Methods Cell Biol 63: 185–204
Jonker A, Geerts WJ, Chieco P, Moorman AF, Lamers WH, Van Noorden CJ (1997). Basic strategies for valid cytometry using image analysis. Histochem J 29: 347–364
Jovov B, Tousson A, Ji HL, Keeton D, Shlyonsky V, Ripoll PJ, Fuller CM, Benos DJ (1999). Regulation of epithelial Na(+) channels by actin in planar lipid bilayers and in the Xenopus oocyte expression system. J Biol Chem 274: 37845–37854
Lippincott-Schwartz J, Patterson GH (2003). Development and use of fluorescent protein markers in living cells. Science 300: 87–91
Model MA, Burkhardt JK (2001). A standard for calibration and shading correction of a fluorescence microscope. Cytometry 44: 309–316
Netten H, Ellenberger SL, Young IT (1996). Illumination calibration for fluorescence microscopy. In International society for analytical cytology. Vol. Supplement 8, Abstract 57. Cytometry, Rimini, Italy. 109
Panchuk-Voloshina N, Haugland RP, Bishop-Stewart J, Bhalgat MK, Millard PJ, Mao F, Leung WY (1999). Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J Histochem Cytochem 47: 1179–1188
Souchier C, Ffrench M, Benchaib M, Catallo R, Bryon PA (1995). Methods for cell proliferation analysis by fluorescent image cytometry. Cytometry 20: 203–209
Vigo J, Salmon JM, Lahmy S, Viallet P (1991). Fluorescent image cytometry: from qualitative to quantitative measurements. Anal Cell Pathol 3: 145–165
Vischer NO, Huls PG, Ghauharali RI, Brakenhoff GJ, Nanninga N, Woldringh CL (1999). Image cytometric method for quantifying the relative amount of DNA in bacterial nucleoids using Escherichia coli. J Microsc 196: 61–68
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Souchier, C., Brisson, C., Batteux, B. et al. Data reproducibility in fluorescence image analysis. Methods Cell Sci 25, 195–200 (2004). https://doi.org/10.1007/s11022-004-2383-4
Revised:
Issue Date:
DOI: https://doi.org/10.1007/s11022-004-2383-4