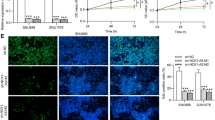
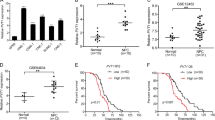
Abstract
Tongue squamous cell carcinoma (TSCC) is prevailing malignancy in the oral and maxillofacial region, characterized by its high frequency. LncRNA CCAT1 can promote tumorigenesis and progression in many cancers. Here, we investigated the regulatory mechanism by which CCAT1 influences growth and metastasis of TSCC. Levels of CCAT1, WTAP, TRIM46, PHLPP2, AKT, p-AKT, and Ki67 in TSCC tissues and cells were assessed utilizing qRT-PCR, Western blot and IHC. Cell proliferation, migration, and invasion were evaluated utilizing CCK8, colony formation, wound healing and transwell assays. Subcellular localization of CCAT1 was detected utilizing FISH assay. m6A level of CCAT1 was assessed using MeRIP. RNA immunoprecipitation (RIP), Co-immunoprecipitation (Co-IP) and RNA pull down elucidated binding relationship between molecules. Nude mouse tumorigenesis experiments were used to verify the TSCC regulatory function of CCAT1 in vivo. Metastatic pulmonary nodules were observed utilizing hematoxylin and eosin (HE) staining. CCAT1 silencing repressed TSCC cell proliferation, migration and invasion. Expression of CCAT1 was enhanced through N6-methyladenosine (m6A) modification of its RNA, facilitated by WTAP. Moreover, IGF2BP1 up-regulated CCAT1 expression by stabilizing its RNA transcript. CCAT1 bond to PHLPP2, inducing its ubiquitination and activating AKT signaling. CCAT1 mediated the ubiquitination and degradation of PHLPP2 by TRIM46, thereby promoting TSCC growth and metastasis. CCAT1/TRIM46/PHLPP2 axis regulated proliferation and invasion of TSCC cells, implying that CCAT1 would be a novel therapeutic target for TSCC patients.







Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DAPI:
-
4,6-Diamidino-2-phenylindole
- CCK-8:
-
Cell counting kit-8
- CST:
-
Cell signaling technology
- Co-IP:
-
Co-immunoprecipitation
- CCAT1:
-
Colon cancer-associated transcript-1
- EMT:
-
Epithelial-mesenchymal transition
- FISH:
-
Fluorescence in situ hybridization
- FoxO1:
-
Forkhead box protein O1
- HE staining:
-
Hematoxylin and eosin staining
- IHC:
-
Immunohistochemistry
- IGF2BP1:
-
Insulin-like growth factor 2 mRNA-binding protein 1
- lncRNAs:
-
Long non-coding RNAs
- MeRIP:
-
Methylated RNA immunoprecipitation
- m6A:
-
N6-methyladenosine
- ANOVA:
-
One-way analysis of variance
- PHLPP:
-
PH domain and leucine rich repeat protein phosphatase
- RIP:
-
RNA immunoprecipitation
- SD:
-
Standard deviation
- TRIM:
-
The E3-ligase tripartite motif
- TSCC:
-
Tongue squamous cell carcinoma
- TRIM46:
-
Tripartite motif containing 46
- WTAP:
-
Wilms tumor 1-associated protein
References
Melo BAC et al (2021) Human papillomavirus infection and oral squamous cell carcinoma - a systematic review. Braz J Otorhinolaryngol 87(3):346–352
Lenze NR et al (2020) Age and risk of recurrence in oral tongue squamous cell carcinoma: systematic review. Head Neck 42(12):3755–3768
Joshi P, Waghmare S (2023) Molecular signaling in cancer stem cells of tongue squamous cell carcinoma: therapeutic implications and challenges. World J Stem Cells 15(5):438–452
Matsuo K et al (2022) Squamous cell carcinoma of the tongue: subtypes and morphological features affecting prognosis. Am J Physiol Cell Physiol 323(6):C1611-c1623
Jarroux J, Morillon A, Pinskaya M (2017) History, discovery, and Classification of lncRNAs. Adv Exp Med Biol 1008:1–46
Chen J et al (2020) Progress in the study of long noncoding RNA in tongue squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 129(1):51–58
Guo X, Hua Y (2017) CCAT1: an oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol 143(4):555–562
Li GH, Ma ZH, Wang X (2019) Long non-coding RNA CCAT1 is a prognostic biomarker for the progression of oral squamous cell carcinoma via miR-181a-mediated Wnt/β-catenin signaling pathway. Cell Cycle 18(21):2902–2913
Sun M, Shen Z (2020) Knockdown of long non-coding RNA (lncRNA) colon cancer-associated transcript-1 (CCAT1) suppresses oral squamous cell carcinoma proliferation, invasion, and migration by inhibiting the discoidin domain receptor 2 (DDR2)/ERK/AKT Axis. Med Sci Monit 26:e920020
Jiang X et al (2021) The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 6(1):74
Zuo X et al (2020) M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol 13(1):5
Rong D et al (2021) m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids 26:637–648
Pan Q et al (2023) WTAP contributes to the tumorigenesis of osteosarcoma via modulating ALB in an m6A-dependent manner. Genomics. https://doi.org/10.1016/j.ygeno.2023.110566
Paramasivam A, George R, Priyadharsini JV (2021) Genomic and transcriptomic alterations in m6A regulatory genes are associated with tumorigenesis and poor prognosis in head and neck squamous cell carcinoma. Am J Cancer Res 11(7):3688–3697
Huang X et al (2018) Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J Hematol Oncol 11(1):88
Cha JH et al (2021) Emerging roles of PHLPP phosphatases in metabolism. BMB Rep 54(9):451–457
Chen S et al (2019) miR-762 promotes malignant development of head and neck squamous cell carcinoma by targeting PHLPP2 and FOXO4. Onco Targets Ther 12:11425–11436
Brognard J et al (2007) PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell 25(6):917–931
Peng M et al (2022) Programmed death-ligand 1 signaling and expression are reversible by lycopene via PI3K/AKT and Raf/MEK/ERK pathways in tongue squamous cell carcinoma. Genes Nutr 17(1):3
Hatakeyama S (2017) TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci 42(4):297–311
Tantai J et al (2022) TRIM46 activates AKT/HK2 signaling by modifying PHLPP2 ubiquitylation to promote glycolysis and chemoresistance of lung cancer cells. Cell Death Dis 13(3):285
Chu Z et al (2021) FOXO3A-induced LINC00926 suppresses breast tumor growth and metastasis through inhibition of PGK1-mediated Warburg effect. Mol Ther 29(9):2737–2753
Hu Y et al (2020) A reciprocal feedback of Myc and lncRNA MTSS1-AS contributes to extracellular acidity-promoted metastasis of pancreatic cancer. Theranostics 10(22):10120–10140
Cusenza VY et al (2023) The lncRNA epigenetics: the significance of m6A and m5C lncRNA modifications in cancer. Front Oncol 13:1063636
Chen C et al (2021) m6A Modification in Non-Coding RNA: The Role in Cancer Drug Resistance. Front Oncol 11:746789
Li CF et al (2017) miR-938 promotes colorectal cancer cell proliferation via targeting tumor suppressor PHLPP2. Eur J Pharmacol 807:168–173
Ding L et al (2017) MicroRNA-27a contributes to the malignant behavior of gastric cancer cells by directly targeting PH domain and leucine-rich repeat protein phosphatase 2. J Exp Clin Cancer Res 36(1):45
Liao Y et al (2016) MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol 143(3):655–663
Kim K et al (2017) Degradation of PHLPP2 by KCTD17, via a glucagon-dependent pathway. Promotes Hepatic Steatosis Gastroenterol 153(6):1568-1580.e10
Liu L et al (2022) MARCH1 silencing suppresses growth of oral squamous cell carcinoma through regulation of PHLPP2. Clin Transl Oncol 24(7):1311–1321
Lin Y et al (2022) A narrative review on machine learning in diagnosis and prognosis prediction for tongue squamous cell carcinoma. Transl Cancer Res 11(12):4409–4415
Mohideen K et al (2019) Meta-analysis on risk factors of squamous cell carcinoma of the tongue in young adults. J Oral Maxillofac Pathol 23(3):450–457
Hu Z, Zhao P, Xu H (2020) Hyperoside exhibits anticancer activity in non-small cell lung cancer cells with T790M mutations by upregulating FoxO1 via CCAT1. Oncol Rep 43(2):617–624
Xing L et al (2019) Silencing FOXO1 attenuates dexamethasone-induced apoptosis in osteoblastic MC3T3-E1 cells. Biochem Biophys Res Commun 513(4):1019–1026
Li X et al (2020) Knockdown of lncRNA CCAT1 enhances sensitivity of paclitaxel in prostate cancer via regulating miR-24-3p and FSCN1. Cancer Biol Ther 21(5):452–462
Mu Y, Li N, Cui YL (2018) The lncRNA CCAT1 upregulates TGFβR1 via sponging miR-490-3p to promote TGFβ1-induced EMT of ovarian cancer cells. Cancer Cell Int 18:145
Yang F et al (2022) LncRNA CCAT1 upregulates ATG5 to enhance autophagy and promote gastric cancer development by absorbing miR-140-3p. Dig Dis Sci 67(8):3725–3741
Wen H et al (2023) m6A modification-mediated BATF2 suppresses metastasis and angiogenesis of tongue squamous cell carcinoma through inhibiting VEGFA. Cell Cycle 22(1):100–116
Fan Y et al (2022) Role of WTAP in cancer: from mechanisms to the therapeutic potential. Biomolecules. https://doi.org/10.3390/biom12091224
Liu F et al (2023) m6A methyltransferase, WTAP, promotes cancer progression in laryngeal squamous cell carcinoma by regulating PLAU stability. Ann Clin Lab Sci 53(2):293–302
Yang H et al (2019) LncRNA THOR promotes tongue squamous cell carcinomas by stabilizing IGF2BP1 downstream targets. Biochimie 165:9–18
Huang H et al (2018) Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20(3):285–295
Müller S et al (2019) IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res 47(1):375–390
Nowak DG et al (2019) The PHLPP2 phosphatase is a druggable driver of prostate cancer progression. J Cell Biol 218(6):1943–1957
Harterink M et al (2019) TRIM46 organizes microtubule fasciculation in the axon initial segment. J Neurosci 39(25):4864–4873
Zhang L et al (2016) Mmu-miR-1894–3p inhibits cell proliferation and migration of breast cancer cells by targeting Trim46. Int J Mol Sci. https://doi.org/10.3390/ijms17040609
Funding
The research was supported by 2021 Doctoral Fund project of Hunan Provincial People’s Hospital [BSJJ202120], Fund project of Hunan Provincial Department of Finance [2022CZT02].
Author information
Authors and Affiliations
Contributions
Feng Liu: Conceptualization; Data Curation; Writing—Original Draft; Funding acquisition. Hanlin Yang: Methodology; Supervision; Xiongwei Liu: Formal analysis; Project administration; Yangbo Ning: Validation; Yiwei Wu: Investigation; Xinglan Yan: Resources; Huixi Zheng: Visualization; Chang Liu: Writing—Review & Editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare that there is no conflict of interest.
Ethical approval and consent to participate
Ethics Committee of Hunan Provincial People’s Hospital (The First Affiliated Hospital of Hunan Normal University) approved all animal experiments conducted in this study [2022–160].
Consent for publication
N/A.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11010_2024_5004_MOESM1_ESM.tif
Supplementary file1 (TIF 8040 KB)— (A) SRAMP predicted potential m6A site of CCAT1. (B) NEDD4, SMURF1 and NEDD4L are potential ubiquitinases of PHLPP2 predicted by UbiBrowser 2.0. (C, D) catRAPID predicted the potential binding relationship between CCAT1 and TRIM46, PHLPP2
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, F., Yang, H., Liu, X. et al. LncRNA CCAT1 knockdown suppresses tongue squamous cell carcinoma progression by inhibiting the ubiquitination of PHLPP2. Mol Cell Biochem (2024). https://doi.org/10.1007/s11010-024-05004-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11010-024-05004-1