Abstract
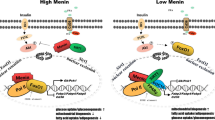
Non-alcoholic fatty liver disease (NAFLD) is rapidly being recognized as the leading cause of chronic liver disease worldwide. Men1, encoding protein of menin, is a key causative gene of multiple endocrine neoplasia type 1 syndrome including pancreatic tumor. It is known that insulin that secretes by endocrine tissue pancreatic islets plays a critical role in hepatic metabolism. Mouse model of hemizygous deletion of Men1 was shown to have severe hepatic metabolism disorders. However, the molecular function of menin on lipid deposition in hepatocytes needs to be further studied. Transcriptome sequencing does show that expression suppression of Men1 in mouse hepatocytes widely affect signaling pathways involved in hepatic metabolism, such as fatty acid metabolism, insulin response, glucose metabolism and inflammation. Further molecular studies indicates that menin overexpression inhibits expressions of the fat synthesis genes Srebp-1c, Fas, and Acc1, the fat differentiation genes Pparγ1 and Pparγ2, and the fat transport gene Cd36, thereby inhibiting the fat accumulation in hepatocytes. The biological process of menin regulating hepatic lipid metabolism was accomplished by interacting with the transcription factor FoxO1, which is also found to be critical for lipid metabolism. Moreover, menin responds to insulin in hepatocytes and mediates its regulatory effect on hepatic metabolism. Our findings suggest that menin is a crucial mediation factor in regulating the hepatic fat deposition, suggesting it could be a potential important therapeutic target for NAFLD.







Similar content being viewed by others
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Abbreviations
- Acc1:
-
Acetyl CoA carboxylase
- Akt, also named as PKB:
-
Protein kinase B
- Cd36:
-
A fatty acid transporter
- FoxO1:
-
Forkhead box O1
- ChIP:
-
Chromatin immunoprecipitation
- DEGs:
-
Differentially expressed genes
- Fas:
-
Fatty acid synthase
- Gk:
-
Glucokinase
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MEN1:
-
Multiple endocrine neoplasia type 1
- NAFLD:
-
Nonalcoholic fatty liver disease
- OA:
-
Oleic acid
- Pparγ:
-
Peroxisome proliferator-activated receptor gamma
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- SIRT1:
-
Sirtuin 1
- Srebp-1c:
-
Sterol regulatory element-binding protein 1C
- T2DM:
-
Type 2 diabetes mellitus
- TG:
-
Triglyceride
References
Younossi ZM (2019) Non-alcoholic fatty liver disease - A global public health perspective. J Hepatol 70:531–544. https://doi.org/10.1016/j.jhep.2018.10.033
Younossi ZM, Loomba R, Rinella ME, Bugianesi E, Marchesini G, Neuschwander-Tetri BA, Serfaty L, Negro F, Caldwell SH, Ratziu V, Corey KE, Friedman SL, Abdelmalek MF, Harrison SA, Sanyal AJ, Lavine JE, Mathurin P, Charlton MR, Chalasani NP, Anstee QM, Kowdley KV, George J, Goodman ZD, Lindor K (2018) Current and future therapeutic regimens for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 68:361–371. https://doi.org/10.1002/hep.29724
Rotman Y, Sanyal AJ (2017) Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut 66:180–190. https://doi.org/10.1136/gutjnl-2016-312431
Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS, Wang Y, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong Q, Spiegel AM, Burns AL, Marx SJ (1997) Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 276:404–407. https://doi.org/10.1126/science.276.5311.404
Song TY, Lim J, Kim B, Han JW, Youn HD, Cho EJ (2014) The role of tumor suppressor menin in IL-6 regulation in mouse islet tumor cells. Biochem Biophys Res Commun 451:308–313. https://doi.org/10.1016/j.bbrc.2014.07.113
Iyer S, Agarwal SK (2018) Epigenetic regulation in the tumorigenesis of MEN1-associated endocrine cell types. J Mol Endocrinol 61:R13-r24. https://doi.org/10.1530/jme-18-0050
Gao SB, Hua X, Jin GH (2008) Menin regulates endocrine diseases by controlling histone modification and gene transcription. Ann Endocrinol (Paris) 69:426–432. https://doi.org/10.1016/j.ando.2008.06.001
Matkar S, Thiel A, Hua X (2013) Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci 38:394–402. https://doi.org/10.1016/j.tibs.2013.05.005
Ehrlich L, Hall C, Meng F, Lairmore T, Alpini G, Glaser S (2017) A review of the scaffold protein menin and its role in hepatobiliary pathology. Gene Expr 17:251–263. https://doi.org/10.3727/105221617x695744
Shi K, Liu X, Li H, Lin X, Yan Z, Cao Q, Zhao M, Xu Z, Wang Z (2017) Menin modulates mammary epithelial cell numbers in bovine mammary glands through cyclin D1. J Mammary Gland Biol Neoplasia 22:221–233. https://doi.org/10.1007/s10911-017-9385-8
Qiaoqiao C, Li H, Liu X, Yan Z, Zhao M, Xu Z, Wang Z, Shi K (2019) MiR-24-3p regulates cell proliferation and milk protein synthesis of mammary epithelial cells through menin in dairy cows. J Cell Physiol 234:1522–1533. https://doi.org/10.1002/jcp.27017
Wuescher L, Angevine K, Patel PR, Mensah-Osman E (2012) Menin liver-specific hemizygous mice challenged with high fat diet show increased weight gain and markers of metabolic impairment. Nutr Diabetes 2:e34. https://doi.org/10.1038/nutd.2012.7
Yang Y, Gurung B, Wu T, Wang H, Stoffers DA, Hua X (2010) Reversal of preexisting hyperglycemia in diabetic mice by acute deletion of the Men1 gene. Proc Natl Acad Sci U S A 107:20358–20363. https://doi.org/10.1073/pnas.1012257107
Schnepp RW, Chen YX, Wang H, Cash T, Silva A, Diehl JA, Brown E, Hua X (2006) Mutation of tumor suppressor gene Men1 acutely enhances proliferation of pancreatic islet cells. Cancer Res 66:5707–5715. https://doi.org/10.1158/0008-5472.can-05-4518
Barbu A, Lejonklou MH, Skogseid B (2016) Progranulin stimulates proliferation of mouse pancreatic islet cells and is overexpressed in the endocrine pancreatic tissue of an MEN1 mouse model. Pancreas 45:533–540. https://doi.org/10.1097/mpa.0000000000000509
Cao Y, Xue Y, Xue L, Jiang X, Wang X, Zhang Z, Yang J, Lu J, Zhang C, Wang W, Ning G (2013) Hepatic menin recruits SIRT1 to control liver steatosis through histone deacetylation. J Hepatol 59:1299–1306. https://doi.org/10.1016/j.jhep.2013.07.011
Wu Y, Pan Q, Yan H, Zhang K, Guo X, Xu Z, Yang W, Qi Y, Guo CA, Hornsby C, Zhang L, Zhou A, Li L, Chen Y, Zhang W, Sun Y, Zheng H, Wondisford F, He L, Guo S (2018) Novel mechanism of Foxo1 phosphorylation in glucagon signaling in control of glucose homeostasis. Diabetes 67:2167–2182. https://doi.org/10.2337/db18-0674
Wuescher L, Angevine K, Hinds T, Ramakrishnan S, Najjar SM, Mensah-Osman EJ (2011) Insulin regulates menin expression, cytoplasmic localization, and interaction with FOXO1. Am J Physiol Endocrinol Metab 301:E474–E483. https://doi.org/10.1152/ajpendo.00022.2011
Yue SJ, Zhao YQ, Gu XR, Yin B, Jiang YL, Wang ZH, Shi KR (2017) A genome-wide association study suggests new candidate genes for milk production traits in Chinese Holstein cattle. Anim Genet 48:677–681. https://doi.org/10.1111/age.12593
Yan Z, Wang Z, Zhang Q, Yue S, Yin B, Jiang Y, Shi K (2020) Identification of whole-genome significant single nucleotide polymorphisms in candidate genes associated with body conformation traits in Chinese Holstein cattle. Anim Genet 51:141–146. https://doi.org/10.1111/age.12865
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297. https://doi.org/10.1093/nar/gkl031
Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M (2006) KEGG as a glycome informatics resource. Glycobiology 16:63r–70r. https://doi.org/10.1093/glycob/cwj010
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
Zhao NQ, Li XY, Wang L, Feng ZL, Li XF, Wen YF, Han JX (2017) Palmitate induces fat accumulation by activating C/EBPβ-mediated G0S2 expression in HepG2 cells. World J Gastroenterol 23:7705–7715. https://doi.org/10.3748/wjg.v23.i43.7705
Yan D, Dou QL, Wang Z, Wei YY (2015) Establishment of a hepatocyte steatosis model using Chang liver cells. Genet Mol Res 14:15224–15232. https://doi.org/10.4238/2015.November.25.10
Rafiei H, Omidian K, Bandy B (2019) Dietary polyphenols protect against oleic acid-induced steatosis in an in vitro model of NAFLD by modulating lipid metabolism and improving mitochondrial function. Nutrients. https://doi.org/10.3390/nu11030541
Engin A (2017) Non-alcoholic fatty liver disease. Adv Exp Med Biol 960:443–467. https://doi.org/10.1007/978-3-319-48382-5_19
Higuchi N, Kato M, Shundo Y, Tajiri H, Tanaka M, Yamashita N, Kohjima M, Kotoh K, Nakamuta M, Takayanagi R, Enjoji M (2008) Liver X receptor in cooperation with SREBP-1c is a major lipid synthesis regulator in nonalcoholic fatty liver disease. Hepatol Res 38:1122–1129. https://doi.org/10.1111/j.1872-034X.2008.00382.x
Luo H, Zhou Y, Hu X, Peng X, Wei H, Peng J, Jiang S (2015) Activation of PPARgamma2 by PPARgamma1 through a functional PPRE in transdifferentiation of myoblasts to adipocytes induced by EPA. Cell Cycle 14:1830–1841. https://doi.org/10.1080/15384101.2015.1033594
Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M (2016) Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64:73–84. https://doi.org/10.1002/hep.28431
Titchenell PM, Quinn WJ, Lu M, Chu Q, Lu W, Li C, Chen H, Monks BR, Chen J, Rabinowitz JD, Birnbaum MJ (2016) Direct hepatocyte insulin signaling is required for lipogenesis but is dispensable for the suppression of glucose production. Cell Metab 23:1154–1166. https://doi.org/10.1016/j.cmet.2016.04.022
Lai S, Zhou H, Xiong W, Gilbert M, Huang Z, Yu J, Yin W, Wang L, Chen Q, Li Y, Mu D, Zeng L, Ren X, Geng M, Zhang Z, Cui B, Li T, Wang D, Li Z, Wardrop NA, Tatem AJ, Yu H (2017) Changing epidemiology of human brucellosis, China, 1955–2014. Emerg Infect Dis 23:184–194. https://doi.org/10.3201/eid2302.151710
Altomonte J, Cong L, Harbaran S, Richter A, Xu J, Meseck M, Dong HH (2004) Foxo1 mediates insulin action on apoC-III and triglyceride metabolism. J Clin Invest 114:1493–1503. https://doi.org/10.1172/jci19992
Qu S, Altomonte J, Perdomo G, He J, Fan Y, Kamagate A, Meseck M, Dong HH (2006) Aberrant Forkhead box O1 function is associated with impaired hepatic metabolism. Endocrinology 147:5641–5652. https://doi.org/10.1210/en.2006-0541
Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG (2006) FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem 281:10105–10117. https://doi.org/10.1074/jbc.M600272200
Ren B, Best B, Ramakrishnan DP, Walcott BP, Storz P, Silverstein RL (2016) LPA/PKD-1-FoxO1 signaling axis mediates endothelial cell CD36 transcriptional repression and proangiogenic and proarteriogenic reprogramming. Arterioscler Thromb Vasc Biol 36:1197–1208. https://doi.org/10.1161/atvbaha.116.307421
Li H, Liu X, Wang Z, Lin X, Yan Z, Cao Q, Zhao M, Shi K (2017) MEN1/Menin regulates milk protein synthesis through mTOR signaling in mammary epithelial cells. Sci Rep 7:5479. https://doi.org/10.1038/s41598-017-06054-w
Os I, Zhang W, Wasserman DH, Liew CW, Liu J, Paik J, DePinho RA, Stolz DB, Kahn CR, Schwartz MW, Unterman TG (2015) FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat Commun 6:7079. https://doi.org/10.1038/ncomms8079
Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ (2018) Mechanisms of NAFLD development and therapeutic strategies. Nat Med 24:908–922. https://doi.org/10.1038/s41591-018-0104-9
Eng JM, Estall JL (2021) Diet-induced models of non-alcoholic fatty liver disease: food for thought on sugar, fat, and cholesterol. Cells. https://doi.org/10.3390/cells10071805
Asgharpour A, Cazanave SC, Pacana T, Seneshaw M, Vincent R, Banini BA, Kumar DP, Daita K, Min HK, Mirshahi F, Bedossa P, Sun X, Hoshida Y, Koduru SV, Contaifer D Jr, Warncke UO, Wijesinghe DS, Sanyal AJ (2016) A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J Hepatol 65:579–588. https://doi.org/10.1016/j.jhep.2016.05.005
Acknowledgements
We are grateful to Prof. Shaorong Gao in Tongji University for his helpful discussions and excellent suggestions for the project and manuscript. We also thank all present and past members of our laboratory who have contributed comments and ideas.
Funding
This work was financially supported by the Natural Science Foundation of Shandong (ZR2020MC166; ZR2013CM013), the National Key Research and Development Program of China (2021YFD1200900), the National Natural Science Foundation of China (31402054), the Key Project of Agricultural Fine Breeding of Shandong Province (2019LZGC011, 2016LZGC030).
Author information
Authors and Affiliations
Contributions
Shi KR, Wang SX; Liu TJ and Du HX participated in the execution of all experiments, analyzed the data, and drafted the manuscript; Liu TJ, Sun LL, Xu ZJ, and Li RR participated in construction of the expression plasmids, data analysis and discussion; Yu Y and Mao YJ participated in experimental design, coordinated the experiments. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest concerning this article.
Ethical approval
All process was completed on the basis of the Guidelines for Care and Use of Laboratory Animals of Shandong Agricultural University and approved by the Animal Ethics Committee of Shandong Agricultural University.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, S., Liu, T., Sun, L. et al. Menin regulates lipid deposition in mouse hepatocytes via interacting with transcription factor FoxO1. Mol Cell Biochem 477, 1555–1568 (2022). https://doi.org/10.1007/s11010-022-04392-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04392-6