Abstract
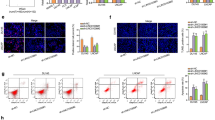
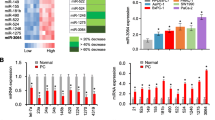
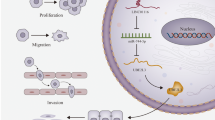
Recent studies have shown that prostate cancer-associated long non-coding RNA, PRNCR1, plays crucial roles in the development of multiple human cancers. However, its role in ovarian cancer is barely known. This study was carried out to investigate the role of PRNCR1 and the underlying mechanisms in OC. The expression of PRNCR1 and miR-653-5p in OC cell lines and tissues were detected by qRT-PCR. The expression of ELF2 protein was evaluated by Western blot analysis. Cell proliferation was measured by colony formation and MTT assay. Cell invasion and migration were evaluated by Transwell and wound healing assay. Luciferase reporter assay and RNA-binding protein immunoprecipitation assay were performed to determine the interaction between miR-653-5p and PRNCR1, as well as between miR-653-5p and ELF2. In vivo tumor xenograft model was established to evaluate the role of PRNCR1 in tumor growth. Our results demonstrated that PRNCR1 was significantly upregulated in both OC cell lines and tissues, and high expression of PRNCR1 was correlated with poor survival of OC patients. Overexpression of PRNCR1 accelerated OC cell invasion, migration and proliferation. Besides, the expression of PRNCR1 was negatively correlated with the expression of miR-653-5p, while positively correlated with the expression of E74-like factor 2 in OC tissues. Importantly, ELF2 could target miR-653-5p, and PRNCR1 increased the expression levels of ELF2 by sponging miR-653-5p in OC cells. Furthermore, the miR-145-5p/ELF2 axis was involved in the regulation of PRNCR1 in OC progression in vivo. PRNCR1 promotes OC tumor progress via the miR-653-5p/ELF2 axis and might be a potential therapeutic target for OC.







Similar content being viewed by others
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to their containing information that could compromise the privacy of research participants.
References
Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, Brawley O, Bray F (2012) International variation in prostate cancer incidence and mortality rates. Eur Urol 61(6):1079–1092. https://doi.org/10.1016/j.eururo.2012.02.054
Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, Stiller C, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC, Coleman MP, Group CW (2015) Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 385 (9972):977–1010. https://doi.org/10.1016/S0140-6736(14)62038-9
Nakamura K, Nakayama K, Ishibashi T, Ishikawa N, Ishikawa M, Katagiri H, Minamoto T, Sato E, Sanuki K, Yamashita H, Iida K, Sultana R, Kyo S (2016) KRAS/BRAF analysis in ovarian low-grade serous carcinoma having synchronous all pathological precursor regions. Int J Mol Sci. https://doi.org/10.3390/ijms17050625
Vang R, Levine DA, Soslow RA, Zaloudek C, Shih Ie M, Kurman RJ (2016) Molecular alterations of TP53 are a defining feature of ovarian high-grade serous carcinoma: a rereview of cases lacking TP53 mutations in the cancer genome atlas ovarian study. Int J Gynecol Pathol 35(1):48–55. https://doi.org/10.1097/PGP.0000000000000207
Pheasant M, Mattick JS (2007) Raising the estimate of functional human sequences. Genome Res 17(9):1245–1253. https://doi.org/10.1101/gr.6406307
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet. https://doi.org/10.1093/hmg/ddl046
Liu C, Bai B, Skogerbo G, Cai L, Deng W, Zhang Y, Bu D, Zhao Y, Chen R (2005) NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. https://doi.org/10.1093/nar/gki041
Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR (2005) Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308(5725):1149–1154. https://doi.org/10.1126/science.1108625
Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143(1):46–58. https://doi.org/10.1016/j.cell.2010.09.001
Lipovich L, Johnson R, Lin CY (2010) MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta 1799(9):597–615. https://doi.org/10.1016/j.bbagrm.2010.10.001
Costa FF (2008) Non-coding RNAs, epigenetics and complexity. Gene 410(1):9–17. https://doi.org/10.1016/j.gene.2007.12.008
Huarte M, Rinn JL (2010) Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19(R2):R152-161. https://doi.org/10.1093/hmg/ddq353
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106(28):11667–11672. https://doi.org/10.1073/pnas.0904715106
Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18(9):1433–1445. https://doi.org/10.1101/gr.078378.108
Gutschner T, Diederichs S (2012) The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 9(6):703–719. https://doi.org/10.4161/rna.20481
Pang D, Hu Q, Lan X, Lin Y, Duan H, Cao S, Lin Y, Li L, Peng F, Pan F (2019) The novel long non-coding RNA PRNCR1-2 is involved in breast cancer cell proliferation, migration, invasion and cell cycle progression. Mol Med Rep 19(3):1824–1832
Cheng D, Bao C, Zhang X, Lin X, Huang H, Zhao L (2018) LncRNA PRNCR1 interacts with HEY2 to abolish miR-448-mediated growth inhibition in non-small cell lung cancer. Biomed Pharmacothera 107:1540–1547
Chu H, Chen Y, Yuan Q, Hua Q, Zhang X, Wang M, Tong N, Zhang W, Chen J, Zhang Z (2017) The HOTAIR, PRNCR1 and POLR2E polymorphisms are associated with cancer risk: a meta-analysis. Oncotarget 8(26):43271–43283
Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li M, Xu L, Yin R (2016) Upregulation of long non-coding RNA PRNCR1 in colorectal cancer promotes cell proliferation and cell cycle progression. Oncology Rep 35(1):318–324
Han W, Wang L, Zhang L, Wang Y, Li Y (2019) Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593-3p/CCND2 and miR-653-5p/TIAM1 pathways in non-small cell lung cancer. Biochem Biophy Res Commun 510(3):462–466
Chi R, Chen X, Liu M, Zhang H, Li F, Fan X, Wang W, Lu H (2019) Role of SNHG7-miR-653-5p-STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol 234(8):13403–13412
Ando M, Kawazu M, Ueno T, Koinuma D, Ando K, Koya J, Kataoka K, Yasuda T, Yamaguchi H, Fukumura K, Yamato A (2016) Mutational landscape and antiproliferative functions of ELF transcription factors in human cancer. Cancer Res 76(7):1814–1824
Liu Y, Tao Z, Qu J, Zhou X, Zhang C (2017) Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochem Biophy Res Commun 491(2):374–381
Zhang J, Hou W, Jia J, Zhao Y, Zhao B (2017) MiR-409-3p regulates cell proliferation and tumor growth by targeting E74-like factor 2 in osteosarcoma. FEBS Open Bio 7(3):348–357
Funding
The study was financially supported by the National Natural Science Foundation of China (Grant Number: 81603138) and the Special Fund for Guiding Local Science and Technology Development awarded by the Central Government of Anhui Province (Grant Number: 2017070802d149).
Author information
Authors and Affiliations
Contributions
XQ, XT: study concepts, literature research, clinical studies, data analysis, experimental studies, manuscript writing and review; DC: study design, literature research, experimental studies and manuscript editing; WY: definition of intellectual content, clinical studies, data acquisition and statistical analysis; LW: data acquisition, manuscript preparation and data analysis; LL: data acquisition and statistical analysis. All authors have read and approve the submission of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All other authors have no conflicts of interest. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Ethical approval
This study was approved and conducted in compliance with the guidelines by the Ethics and Scientific Committee of Hubei Provincial Maternal and Child Health Hospital, Tongji Medical College, Huazhong University of Science and Technology and the research has been carried out in accordance with the World Medical Association Declaration of Helsinki.
Consent to participate
Written informed consent was acquired from all enrolled patients.
Consent to publish
Not applicable.
Research involving animal rights
All animal experiments were approved by the Animal Research Ethics Committee of Hubei Provincial Maternal and Child Health Hospital, Tongji Medical College, Huazhong University of Science and Technology, and were performed according to the Guide for the Care and Use of Laboratory Animals published by the National Research Council in China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qi, X., Chen, D., Yu, W. et al. Long non-coding RNA PRNCR1 promotes ovarian cancer cell proliferation, migration and invasion by targeting the miR-653-5p/ELF2 axis. Mol Cell Biochem 477, 1463–1475 (2022). https://doi.org/10.1007/s11010-022-04371-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04371-x