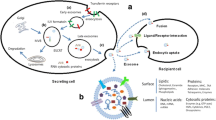
Abstract
Blood exosomes help regulate communication between tumour cells, moderating their behaviour. We sought to determine the protein content in serum exosomes (SEs), to characterise SEs, and to discover novel clinical biomarkers of oral squamous cell carcinoma (OSCC). Differentially expressed proteins (DEPs) of OSCC were identified using proteomics and then analysed using bioinformatics, before validation using ELISA, IHC, and RT-PCR. The influence of SEs on oral cancer cells was detected using CCK-8 and migration assays. Twelve DEPs were found in SEs from OSCC. Four proteins were targeted for further verification. New biomarkers exhibiting high sensitivity and specificity in diagnosing OSCC comprised C-reactive protein (CRP), von willebrand factor (VWF), and leucine-rich alpha-2-glycoprotein (LRG). Combined biomarkers outperformed any single protein. We also demonstrated that tumour-derived exosomes promoted tumour cell migration, but not proliferation and apoptosis. Our study indicates that CRP, VWF, and LRG are potential clinically relevant OSCC biomarkers. OSCC-related SEs may help promote migration of oral cells.






Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- OSCC:
-
Oral squamous cell carcinomas
- DEPs:
-
Differential expressed proteins
- CRP:
-
C-reactive protein
- VWF:
-
Von Willebrand factor
- LRG1:
-
Leucine-rich alpha-2-glycoprotein
- OC:
-
Oral cancer
- HCs:
-
Healthy controls
- MS:
-
Mass spectrometry
- GO:
-
Gene ontology
- PPI network:
-
Protein interaction network
- STRING:
-
Search tool for the retrieval of interacting genes
- KEGG:
-
Kyoto encyclopedia of genes and genomes pathways
- LNM:
-
Lymph node metastasis
- NLNM:
-
No lymph node metastasis
- C1QA:
-
Complement C1q subcomponent subunit A
- GEPIA:
-
Gene expression profiling interactive analysis
- ROCs:
-
Receiver operation characteristic curves
- AUC:
-
The area under the curve
- CCK-8:
-
Cell Counting Kit-8
- PI:
-
Propidium iodide
- BP:
-
Biological processes
- CC:
-
Cellular component
- MF:
-
Molecular functions
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386
Chi AC, Day TA, Neville BW (2015) Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin 65:401–421
Argiris A, Karamouzis MV, Raben D, Ferris RL (2008) Head and neck cancer. Lancet 371:1695–1709
Dionne KR, Warnakulasuriya S, Zain RB, Cheong SC (2015) Potentially malignant disorders of the oral cavity: current practice and future directions in the clinic and laboratory. Int J Cancer 136:503–515
Speight PM, Khurram SA, Kujan O (2018) Oral potentially malignant disorders: risk of progression to malignancy. Oral Surg Oral Med Oral Pathol Oral Radiol 125:612–627
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A (2017) Colorectal cancer statistics, 2017. CA Cancer J Clin 67:177–193
Macey R, Walsh T, Brocklehurst P, Kerr AR, Liu JL, Lingen MW, Ogden GR, Warnakulasuriya S, Scully C (2015) Diagnostic tests for oral cancer and potentially malignant disorders in patients presenting with clinically evident lesions. Cochrane Database Syst Rev. https://doi.org/10.1002/14651858.CD010276.pub2
Jiao YJ, Jin DD, Jiang F, Liu JX, Qu LS, Ni WK, Liu ZX, Lu CH, Ni RZ, Zhu J (2019) Characterization and proteomic profiling of pancreatic cancer-derived serum exosomes. J Cell Biochem 120:988–999
Wang L, Li Y, Guan X, Zhao J, Shen L, Liu J (2018) Exosomal double-stranded DNA as a biomarker for the diagnosis and preoperative assessment of pheochromocytoma and paraganglioma. Mol Cancer 17:128
Li S, Zhao Y, Chen W, Yin L, Zhu J, Zhang H, Cai C, Li P, Huang L, Ma P (2018) Exosomal ephrinA2 derived from serum as a potential biomarker for prostate cancer. J Cancer 9:2659–2665
Sun B, Li Y, Zhou Y, Ng TK, Zhao C, Gan Q, Gu X, Xiang J (2019) Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol 234:1416–1425
Sandhu C, Qureshi A, Emili A (2018) Panomics for precision medicine. Trends Mol Med 24(1):85–101
Fu H, Yang H, Zhang X, Wang B, Mao J, Li X, Wang M, Zhang B, Sun Z, Qian H (2018) Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J Exp Clin Cancer Res 37:162
Andaloussi SEL, Mager I, Breakefield XO, Wood MJ (2013) Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 12:347–57
Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N (2015) Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523:177–182
Tkach M, Thery C (2016) Communication by extracellular vesicles: where we are and where we need to go. Cell 164:1226–1232
Colombo M, Raposo G, Thery C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289
Saleem SN, Abdel-Mageed AB (2015) Tumor-derived exosomes in oncogenic reprogramming and cancer progression. Cell Mol Life Sci 72:1–10
Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J (2014) Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 5:5439–5452
Zhao L, Liu W, Xiao J, Cao B (2015) The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett 356:339–346
Kahlert C, Kalluri R (2013) Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med 91:431–7
Yang C, Robbins PD (2011) The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol 2011:1–11
Mrizak D, Martin N, Barjon C, Jimenez-Pailhes AS, Mustapha R, Niki T, Guigay J, Pancre V, de Launoit Y, Busson P (2015) Effect of nasopharyngeal carcinoma-derived exosomes on human regulatory T cells. J Natl Cancer Inst 107:363
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361
Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S (2009) An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics 8:1891–1907
Pang CY, Wang H, Pang Y, Xu C, Jiao Y, Qin YM, Western TL, Yu SX, Zhu YX (2010) Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and arabidopsis root hair elongation. Mol Cell Proteomics 9:2019–2033
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845
Li C, Zhou Y, Liu J (2019) Potential markers from serum-purified exosomes for detecting oral squamous cell carcinoma metastasis. Cancer Epidemiol Biomarkers Prev 28(10):1668–1681
Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, Li X, Chen J, Liu K, Zhu G (2016) Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res 76:1770–1780
Sento S, Sasabe E, Yamamoto T (2016) Application of a persistent heparin treatment inhibits the malignant potential of oral squamous carcinoma cells induced by tumor cell-derived exosomes. PLoS One 11:e0148454
Sakha S, Muramatsu T, Ueda K, Inazawa J (2016) Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep 6:38750
Chen Y, Xie Y, Xu L, Zhan S, Xiao Y, Gao Y, Wu B, Ge W (2017) Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer 140:900–913
Heikkila K, Ebrahim S, Lawlor DA (2007) A systematic review of the association between circulating concentrations of C reactive protein and cancer. J Epidemiol Commun Health 61:824–833
Acharya S, Kale J, Hallikeri K, Anehosur V, Arnold D (2018) Clinical significance of preoperative serum C-reactive protein in oral squamous cell carcinoma. Int J Oral Maxillofac Surg 47:16–23
Hsu YP, Hsieh CH, Chien HT, Lai CH, Tsao CK, Liao CT, Kang CJ, Wang HM, Chang JT, Huang SF (2015) Serum markers of CYFRA 21–1 and C-reactive proteins in oral squamous cell carcinoma. World J Surg Oncol 13:253
Xie ZB, Zhang YF, Jin C, Mao YS, Fu DL (2019) LRG-1 promotes pancreatic cancer growth and metastasis via modulation of the EGFR/p38 signaling. J Exp Clin Cancer Res 38:75
Honda H, Fujimoto M, Serada S, Urushima H, Mishima T, Lee H, Ohkawara T, Kohno N, Hattori N, Yokoyama A (2017) Leucine-rich alpha-2 glycoprotein promotes lung fibrosis by modulating TGF-beta signaling in fibroblasts. Physiol Rep 5:e13556
Shinozaki E, Tanabe K, Akiyoshi T, Tsuchida T, Miyazaki Y, Kojima N, Igarashi M, Ueno M, Suenaga M, Mizunuma N (2018) Serum leucine-rich alpha-2-glycoprotein-1 with fucosylated triantennary N-glycan: a novel colorectal cancer marker. BMC Cancer 18:406
Kawahara R, Bollinger JG, Rivera C, Ribeiro AC, Brandao TB, Paes Leme AF, MacCoss MJ (2016) A targeted proteomic strategy for the measurement of oral cancer candidate biomarkers in human saliva. Proteomics 16:159–173
Chen Y, Azman SN, Kerishnan JP, Zain RB, Chen YN, Wong YL, Gopinath SC (2014) Identification of host-immune response protein candidates in the sera of human oral squamous cell carcinoma patients. PLoS One 9:e109012
Sadler JE (2013) von Willebrand factor in its native environment. Blood 121:2583–2584
Yang AJ, Wang M, Wang Y, Cai W, Li Q, Zhao TT, Zhang LH, Houck K, Chen X, Jin YL (2018) Cancer cell-derived von Willebrand factor enhanced metastasis of gastric adenocarcinoma. Oncogenesis 7:12
Pepin M, Kleinjan A, Hajage D, Buller HR, Di Nisio M, Kamphuisen PW, Salomon L, Veyradier A, Stepanian A, Mahe I (2016) ADAMTS-13 and von Willebrand factor predict venous thromboembolism in patients with cancer. J Thromb Haemost 14:306–315
Liu Y, Wang X, Li S, Hu H, Zhang D, Hu P, Yang Y, Ren H (2014) The role of von Willebrand factor as a biomarker of tumor development in hepatitis B virus-associated human hepatocellular carcinoma: a quantitative proteomic based study. J Proteomics 106:99–112
Acknowledgements
The authors would like to thank all the reviewers who participated in the review and MJEditor (www.mjeditor.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work is supported by grants from Guangxi medical and health appropriate technology research and development project (Grant Nos.: S201538), Guangxi Clinical Research Center for Craniofacial Deformity (Grant Nos.: GKAD17129004) and National Natural Science Foundation of China (81670970, 81870748).
Author information
Authors and Affiliations
Contributions
HJG, CPL-collected serum samples; HJG, WDJ, SHH-conducted mass spectrometry analysis, verification tests and statistical analysis; HJG, CPL, XPH-wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of Guangxi Medical University (the committee’s reference number: 2018-082). Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11010_2021_4254_MOESM1_ESM.tif
Supplementary file1: Mass spectra of differentially-expressed proteins. The identified peptide fragments of four biomarkers including the b-ion and y-ion series. A CRP; B LRG; C VWF; D C1QA (TIF 731 kb)
11010_2021_4254_MOESM2_ESM.tif
Supplementary file2: RT-PCR validation of MS/MS data. A1-A2/B1-B2/C1-C2/D1-D2 CRP/LRG/VWF/C1QA mRNA expression levels in tissues assayed by RT-PCR. LNM: oral squamous cell carcinoma with lymph node metastasis; NLNM: oral squamous cell carcinoma with no lymph node metastasis; CT: cancerous tissues; CAT: corresponding adjacent tissues. Data were shown as mean ± SD. (nonparametric test, p < 0 .05 was considered statistically significant) (TIF 25514 kb)
11010_2021_4254_MOESM3_ESM.tif
Supplementary file3: Clinical interrelation from RT-PCR results of tissue samples. A1-A7, B1-B7, C1-C7, D1-D7 represented the expressions of CRP, LRG, VWF, C1QA in gender, nationality, stage, differentiation degree, smoking, drinking, and the grades of age brackets. WDSCC: Well differentiated squamous-cell carcinoma; LDSCC: Low differentiated squamous-cell carcinoma. Data were shown as mean ± SD. (Chi-square test, McNemar and Fisher’s exact tests, p < 0.05 was considered statistically significant) (TIF 25515 kb)
11010_2021_4254_MOESM4_ESM.tif
Supplementary file4: The results from GEPIA analysis. Gene expression profile, stage plots, and survival plots showed the connection between CRP, LRG, VWF, and C1QA and head and neck squamous cell carcinoma. A CRP; B LRG; C VWF; D C1QA (TIF 61956 kb)
Rights and permissions
About this article
Cite this article
Guo, H., Jiang, W., Huang, S. et al. Serum exosome-derived biomarkers for the early detection of oral squamous cell carcinoma. Mol Cell Biochem 476, 4435–4447 (2021). https://doi.org/10.1007/s11010-021-04254-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04254-7