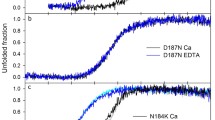
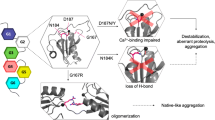
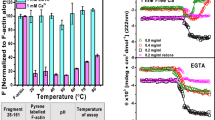
Abstract
Gelsolin, an actin-binding protein, is localized intra- and extracellularly in the bloodstream and throughout the body. Gelsolin amyloidosis is a disease characterized by several point mutations that lead to cleavage and fibrillization of gelsolin. The D187 mutation to N or Y leads to aggregation of peptide fragments with shortest aggregating peptide identified as 182SFNNGDCFILD192. Recently, G167 has also been identified as relevant gelsolin mutation, which leads to gelsolin deposits in kidneys, but its aggregation is much less understood. Hence, we systematically investigated in vitro the aggregation propensities of the following gelsolin peptides: 167GRRVV171 (1), 161RLFQVKG167 (2), 184NNGDCFILDL193 (3), 188CFILDL193 (4), 187DCFILDL193 (5), and their respective mutants (G167K, G167R, N184K, D187Y, D187N), by using spectroscopic methods [fluorescence Proteostat, Thioflavin T (ThT), turbidity assay, and Dynamic Light Scattering (DLS)], and Transmission Electron Microscopy (TEM). The (non) mutant peptides containing CFILDL sequence aggregated into fibrillar networks, while G167R mutation promoted aggregation compared to the wild-type sequence. In the presence of inhibitors, Methylene Blue (MB) and epigallocatechin gallate (EGCG), the gelsolin peptide (3–5) aggregation was reduced with the IC50 values in the 2–13 µM range. We discovered that inhibitors have dual functionality, as aggregation inhibitors and disaggregation promoters, potentially allowing for the prevention and reversal of gelsolin amyloidosis. Such therapeutic strategies may improve outcomes related to other amyloidogenic diseases of the heart, brain, and eye.










Similar content being viewed by others
Data availability
Data are available and included as electronic supplementary material.
References
Solomon JP, Bourgault S, Powers ET, Kelly JW (2011) Heparin binds 8 kDa gelsolin cross-β-sheet oligomers and accelerates amyloidogenesis by hastening fibril extension. Biochemistry 50:2486–2498. https://doi.org/10.1021/bi101905n
Solomon JP, Page LJ, Balch WE, Kelly JW (2013) Gelsolin amyloidosis: genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit Rev Biochem Mol Biol 47:282–296. https://doi.org/10.3109/10409238.2012.661401
Nag S, Ma Q, Wang H et al (2009) Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin. Proc Natl Acad Sci USA 106:13713–13718. https://doi.org/10.1073/pnas.0812374106
Pihlamaa T, Rautio J, Kiuru-Enari S, Suominen S (2011) Gelsolin amyloidosis as a cause of early aging and progressive bilateral facial paralysis. Plast Reconstr Surg 127:2342–2351. https://doi.org/10.1097/PRS.0b013e318213a0a2
Yin HL, Stossel TP (1979) Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature 281:583–586. https://doi.org/10.1038/281583a0
Bonì F, Milani M, Barbiroli A et al (2018) Gelsolin pathogenic Gly167Arg mutation promotes domain-swap dimerization of the protein. Hum Mol Genet 27:53–65. https://doi.org/10.1093/hmg/ddx383
Solomon JP, Yonemoto IT, Murray AN et al (2009) The 8 and 5 kDa fragments of plasma gelsolin form amyloid fibrils by a nucleated polymerization mechanism, while the 68 kDa fragment is not amyloidogenic. Biochemistry 48:11370–11380. https://doi.org/10.1021/bi901368e
Ratnaswamy G, Koepf E, Bekele H et al (1999) The amyloidogenicity of gelsolin is controlled by proteolysis and pH. Chem Biol 6:293–304. https://doi.org/10.1016/S1074-5521(99)80075-1
Arya P, Srivastava A, Vasaikar SV et al (2014) Selective interception of gelsolin amyloidogenic stretch results in conformationally distinct aggregates with reduced toxicity. ACS Chem Neurosci 5:982–992. https://doi.org/10.1021/cn500002v
Srivastava A, Arya P, Goel S et al (2015) Gelsolin amyloidogenesis is effectively modulated by curcumin and emetine conjugated PLGA nanoparticles. PLoS ONE 10:1–18. https://doi.org/10.1371/journal.pone.0127011
Maury CPJ, Nurmiaho-Lassila EL, Boysen G, Liljeström M (2003) Fibrillogenesis in gelsolin-related familial amyloidosis. Amyloid 10:21–25. https://doi.org/10.1080/13506129.2003.12088564
Gazit E (2002) A possible role for π-stacking in the self-assembly of amyloid fibrils. FASEB J 16:77–83. https://doi.org/10.1096/fj.01-0442hyp
Maury CPJ, Nurmiaho-Lassila EL (1992) Creation of amyloid fibrils from mutant Asn187 gelsolin peptides. Biochem Biophys Res Commun 183:227–231. https://doi.org/10.1016/0006-291X(92)91632-Z
Maury CPJ, Liljeström M, Boysen G et al (2000) Danish type gelsolin related amyloidosis: 654G-T mutation is associated with a disease pathogenetically and clinically similar to that caused by the 654G-A mutation (familial amyloidosis of the Finnish type). J Clin Pathol 53:95–99. https://doi.org/10.1136/jcp.53.2.95
Bonì F, Milani M, Porcari R et al (2016) Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant. Sci Rep 6:1–11. https://doi.org/10.1038/srep33463
Sethi S, Theis JD, Quint P et al (2013) Renal amyloidosis associated with a novel sequence variant of gelsolin. Am J Kidney Dis 61:161–166. https://doi.org/10.1053/j.ajkd.2012.07.016
Efebera YA, Sturm A, Baack EC et al (2014) Novel gelsolin variant as the cause of nephrotic syndrome and renal amyloidosis in a large kindred. Amyloid 21:110–112. https://doi.org/10.3109/13506129.2014.891502
Goyal D, Shuaib S, Mann S, Goyal B (2017) Rationally designed peptides and peptidomimetics as inhibitors of amyloid-β (Aβ) aggregation: potential therapeutics of Alzheimer’s disease. ACS Comb Sci 19:55–80. https://doi.org/10.1021/acscombsci.6b00116
Al-Hilaly YK, Pollack SJ, Rickard JE et al (2018) Cysteine-independent inhibition of Alzheimer’s disease-like paired helical filament assembly by leuco-methylthioninium (LMT). J Mol Biol 430:4119–4131. https://doi.org/10.1016/j.jmb.2018.08.010
Necula M, Breydo L, Milton S et al (2007) Methylene blue inhibits amyloid Aβ oligomerization by promoting fibrillization. Biochemistry 46:8850–8860. https://doi.org/10.1021/bi700411k
Cisek K, Cooper GL, Huseby CJ, Kuret J (2014) Structure and mechanism of action of tau aggregation inhibitors. Curr Alzheimer Res 11:918–927. https://doi.org/10.2174/1567205011666141107150331
Reddy S, Aggarwal BB (1994) Curcumin is a non-competitive and selective inhibitor of phosphorylase kinase. FEBS Lett 341:19–22. https://doi.org/10.1016/0014-5793(94)80232-7
Van Overbeke W, Wongsantichon J, Everaert I et al (2015) An ER-directed gelsolin nanobody targets the first step in amyloid formation in a gelsolin amyloidosis mouse model. Hum Mol Genet 24:2492–2507. https://doi.org/10.1093/hmg/ddv010
Jiang D, Rauda I, Han S et al (2012) Aggregation pathways of the amyloid β(1–42) peptide depend on its colloidal stability and ordered β- sheet stacking. Langmuir 28:12711–21721. https://doi.org/10.1021/la3021436
Shi Q, Zhou Y, Sun Y (2005) Influence of pH and ionic strength on the steric mass-action model parameters around the isoelectric point of protein. Biotechnol Prog 21:516–523. https://doi.org/10.1021/bp049735o
Biancalana M, Koide S (2010) Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta - Proteins Proteom 1804:1405–1412. https://doi.org/10.1016/j.bbapap.2010.04.001
De Baets G, Van Doorn L, Rousseau F, Schymkowitz J (2015) Increased aggregation is more frequently associated to human disease-associated mutations than to neutral polymorphisms. PLoS Comput Biol 11:1–14. https://doi.org/10.1371/journal.pcbi.1004374
Close W, Neumann M, Schmidt A et al (2018) Physical basis of amyloid fibril polymorphism. Nat Commun 9:1–7. https://doi.org/10.1038/s41467-018-03164-5
Santhoshkumar P, Sharma KK (2004) Inhibition of amyloid fibrillogenesis and toxicity by a peptide chaperone. Mol Cell Biochem 267:147–155. https://doi.org/10.1023/B:MCBI.0000049373.15558.b8
Novo M, Freire S, Al-Soufi W (2018) Critical aggregation concentration for the formation of early amyloid-β (1–42) oligomers. Sci Rep 8:3–10. https://doi.org/10.1038/s41598-018-19961-3
Santhoshkumar P, Raju M, Sharma KK (2011) αA-crystallin peptide 66SDRDKFVIFLDVKHF80 accumulating in aging lens impairs the function of α-crystallin and induces lens protein aggregation. PLoS ONE. https://doi.org/10.1371/journal.pone.0019291
Zapadka KL, Becher FJ, Gomes dos Santos AL, Jackson SE (2017) Factors affecting the physical stability (aggregation) of peptide therapeutics. Interface Focus. https://doi.org/10.1098/rsfs.2017.0030
Shen C, Murphy RM (1995) Solvent effects on self-assembly of beta-amyloid peptide. Biophys J 69:640–665. https://doi.org/10.1016/S0006-3495(95)79940-4
Tjernberg A, Markova N, Griffiths WJ, Hallén D (2006) DMSO-related effects in protein characterization. J Biomol Screen 11:131–137. https://doi.org/10.1177/1087057105284218
Lesné S, Ming TK, Kotilinek L et al (2006) A specific amyloid-β protein assembly in the brain impairs memory. Nature 440:352–357. https://doi.org/10.1038/nature04533
Shi Y, Mowery RA, Ashley J et al (2012) Abnormal SDS-PAGE migration of cytosolic proteins can identify domains and mechanisms that control surfactant binding. Protein Sci 21:1197–1209. https://doi.org/10.1002/pro.2107
Sengupta U, Nilson AN, Kayed R (2016) the role of amyloid-β oligomers in toxicity, propagation, and immunotherapy. EBioMedicine 6:42–49. https://doi.org/10.1016/j.ebiom.2016.03.035
Streets AM, Sourigues Y, Kopito RR et al (2013) Simultaneous measurement of amyloid fibril formation by dynamic light scattering and fluorescence reveals complex aggregation kinetics. PLoS ONE 8:1–10. https://doi.org/10.1371/journal.pone.0054541
Dahlgren KN, Manelli AM, Blaine Stine W et al (2002) Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J Biol Chem 277:32046–32053. https://doi.org/10.1074/jbc.M201750200
Kumar SS, Singh A, Prakash V, Kumar CS (2014) Structure modeling and dynamics driven mutation and phosphorylation analysis of Beta-amyloid peptides. Bioinformation 10:569–574. https://doi.org/10.6026/97320630010569
Crowe A, James MJ, Virginia MYL et al (2013) Aminothienopyridazines and methylene blue affect tau fibrillization via cysteine oxidation. J Biol Chem 288:11024–11037. https://doi.org/10.1074/jbc.M112.436006
Liu H, Yu L, Dong X, Sun Y (2017) Synergistic effects of negatively charged hydrophobic nanoparticles and (−)-epigallocatechin-3-gallate on inhibiting amyloid β-protein aggregation. J Colloid Interface Sci 491:305–312. https://doi.org/10.1016/j.jcis.2016.12.038
Palhano FL, Lee J, Grimster NP, Kelly JW (2013) Toward the molecular mechanism(s) by which EGCG treatment remodels mature amyloid fibrils. J Am Chem Soc 135:7503–7510. https://doi.org/10.1021/ja3115696
Wobsta HJ, Sharmad A, Diamondd MI et al (2015) The green tea polyphenol (−)-epigallocatechin gallate prevents the aggregation of tau protein into toxic oligomers at substoichiometric ratios. FEBS Lett 589:77–83. https://doi.org/10.1016/j.febslet.2014.11.026
Soeda Y, Yoshikawa M, Almeida OFX et al (2015) Toxic tau oligomer formation blocked by capping of cysteine residues with 1,2-dihydroxybenzene groups. Nat Commun. https://doi.org/10.1038/ncomms10216
Wang J, Ho L, Zhao W et al (2008) Grape-derived polyphenolics prevent Aβ oligomerization and attenuate cognitive deterioration in a mouse model of Alzheimer’s disease. J Neurosci 28:6388–6392. https://doi.org/10.1523/JNEUROSCI.0364-08.2008
Guéroux M, Laguerre M, Szlosek-Pinaud M et al (2012) Polyphenols and alzheimer’s disease: tau/polyphenol interactions investigated by NMR and molecular modelling. Nutr Aging 1:201–206. https://doi.org/10.3233/NUA-130015
Acknowledgements
Mohanad Ahmad thanks the Department of Chemistry at Oakland University for support. Shivani Dabadi carried out preliminary experiments with gelsolin peptide 5. Authors acknowledge Imaging Facilities at Michigan State University and Trent University, Drs. D. Szlag (Oakland University), N. Beyeh (Oakland University), and C. Metcalfe (Trent University) for instrument access. M.A., J. E., C.G., S.M., and C.W. designed the experiments discussed in this study and wrote the manuscript. M.A., J. E., and C. G. carried out the experiments. All authors have given approval to the final version of the manuscript. This work was supported by American Heart Association Grant #18IPA34170477 to S. M. (2018–2020) and G.W. (2019–2020).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahmad, M., Esposto, J., Golec, C. et al. Aggregation of gelsolin wild-type and G167K/R, N184K, and D187N/Y mutant peptides and inhibition. Mol Cell Biochem 476, 2393–2408 (2021). https://doi.org/10.1007/s11010-021-04085-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04085-6