Abstract
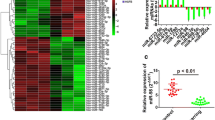
Hypertrophic scar (HS) is a severe skin disorder characterized by excessive extracellular matrix production and abnormal function of fibroblasts. Recent studies have demonstrated that microRNAs (miRNAs) play critical roles in HS formation. This study aims to investigate the role of miR-3613-3p in the formation of HS. The mRNA and miRNA levels were measured by quantitative RT-PCR analysis. The protein levels were examined by Western blot assay. Cell proliferation was determined by Cell Counting Kit-8 assay. The Caspase-3 and Caspase-9 activities were measured using flow cytometry assay. Dual-luciferase activity reporter assay and mRNA-miRNA pulldown assay were conducted to validate the target of miR-3613-3p. miR-3613-3p was downregulated, while arginine and glutamate-rich 1 (ARGLU1) was upregulated in HS fibroblasts (HSFs) and tissues. Overexpression of miR-3613-3p or knockdown of ARGLU1 markedly inhibited the expression of extracellular matrix (ECM) production-associated proteins and promoted Caspase-3 and Caspase-9 activations in HSFs. ARGLU1 was further identified as a direct target of miR-3613-3p. Restoration of ARGLU1 abrogated the suppressive effect of miR-3613-3p on cell proliferation and ECM protein expression of HSFs. Our results demonstrated that miR-3613-3p inhibited HS formation via targeting ARGLU1, which may provide potential therapeutic targets for the management of HS.






Similar content being viewed by others
References
Rabello FB, Souza CD, Farina Junior JA (2014) Update on hypertrophic scar treatment Clinics (Sao Paulo) 69:565–573
Huang C, Murphy GF, Akaishi S, Ogawa R (2013) Keloids and hypertrophic scars: update and future directions. Plast Reconstr Surg Glob Open 1:e25. https://doi.org/10.1097/GOX.0b013e31829c4597
Gauglitz GG, Korting HC, Pavicic T, Ruzicka T, Jeschke MG (2011) Hypertrophic scarring and keloids: pathomechanisms and current and emerging treatment strategies. Mol Med 17:113–125. https://doi.org/10.2119/molmed.2009.00153
Zhang K, Garner W, Cohen L, Rodriguez J, Phan S (1995) Increased types I and III collagen and transforming growth factor-beta 1 mRNA and protein in hypertrophic burn scar. J Invest Dermatol 104:750–754. https://doi.org/10.1111/1523-1747.ep12606979
Slemp AE, Kirschner RE (2006) Keloids and scars: a review of keloids and scars, their pathogenesis, risk factors, and management. Curr Opin Pediatr 18:396–402. https://doi.org/10.1097/01.mop.0000236389.41462.ef
Eming SA, Martin P, Tomic-Canic M (2014) Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 6:265sr6. https://doi.org/10.1126/scitranslmed.3009337
Brown JJ, Bayat A (2009) Genetic susceptibility to raised dermal scarring. Br J Dermatol 161:8–18. https://doi.org/10.1111/j.1365-2133.2009.09258.x
Lee HJ, Jang YJ (2018) Recent understandings of biology, prophylaxis and treatment strategies for hypertrophic scars and keloids. Int J Mol Sci. https://doi.org/10.3390/ijms19030711
Gabriel V (2011) Hypertrophic scar. Phys Med Rehabil Clin N Am 22(301–10):vi. https://doi.org/10.1016/j.pmr.2011.02.002
Ladak A, Tredget EE (2009) Pathophysiology and management of the burn scar. Clin Plast Surg 36:661–674. https://doi.org/10.1016/j.cps.2009.05.014
Finnerty CC, Jeschke MG, Branski LK, Barret JP, Dziewulski P, Herndon DN (2016) Hypertrophic scarring: the greatest unmet challenge after burn injury. Lancet 388:1427–1436. https://doi.org/10.1016/S0140-6736(16)31406-4
Aarabi S, Longaker MT, Gurtner GC (2007) Hypertrophic scar formation following burns and trauma: new approaches to treatment. PLoS Med 4:e234. https://doi.org/10.1371/journal.pmed.0040234
Visscher MO, Bailey JK, Hom DB (2014) Scar treatment variations by skin type. Facial Plast Surg Clin North Am 22:453–462. https://doi.org/10.1016/j.fsc.2014.04.010
O’Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9:402. https://doi.org/10.3389/fendo.2018.00402
Gebert LFR, MacRae IJ (2019) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 20:21–37. https://doi.org/10.1038/s41580-018-0045-7
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. https://doi.org/10.1038/nrm3838
Li Y, Zhang J, Lei Y, Lyu L, Zuo R, Chen T (2017) MicroRNA-21 in skin fibrosis: potential for diagnosis and treatment. Mol Diagn Ther 21:633–642. https://doi.org/10.1007/s40291-017-0294-8
Chen L, Li J, Li Q, Yan H, Zhou B, Gao Y, Li J (2017) Non-coding RNAs: the new insight on hypertrophic scar. J Cell Biochem 118:1965–1968. https://doi.org/10.1002/jcb.25873
Wang J, Zhang Y, Zhang N, Wang C, Herrler T, Li Q (2015) An updated review of mechanotransduction in skin disorders: transcriptional regulators, ion channels, and microRNAs. Cell Mol Life Sci 72:2091–2106. https://doi.org/10.1007/s00018-015-1853-y
Wang X, Zhang Y, Jiang BH, Zhang Q, Zhou RP, Zhang L, Wang C (2017) Study on the role of Hsa-miR-31-5p in hypertrophic scar formation and the mechanism. Exp Cell Res 361:201–209. https://doi.org/10.1016/j.yexcr.2017.09.009
Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J (2011) miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 45:287–294. https://doi.org/10.1165/rcmb.2010-0323OC
Wang L, Zhou L, Jiang P, Lu L, Chen X, Lan H, Guttridge DC, Sun H, Wang H (2012) Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol Ther 20:1222–1233. https://doi.org/10.1038/mt.2012.35
Zhang Y, Huang XR, Wei LH, Chung AC, Yu CM, Lan HY (2014) miR-29b as a therapeutic agent for angiotensin II-induced cardiac fibrosis by targeting TGF-beta/Smad3 signaling. Mol Ther 22:974–985. https://doi.org/10.1038/mt.2014.25
Zhang Q, Guo B, Hui Q, Chang P, Tao K (2018) miR-137 inhibits proliferation and metastasis of hypertrophic scar fibroblasts via targeting pleiotrophin. Cell Physiol Biochem 49:985–995. https://doi.org/10.1159/000493236
Zanotti S, Gibertini S, Curcio M, Savadori P, Pasanisi B, Morandi L, Cornelio F, Mantegazza R, Mora M (2015) Opposing roles of miR-21 and miR-29 in the progression of fibrosis in Duchenne muscular dystrophy. Biochim Biophys Acta 1852:1451–1464. https://doi.org/10.1016/j.bbadis.2015.04.013
Glowacki F, Savary G, Gnemmi V, Buob D, Van der Hauwaert C, Lo-Guidice JM, Bouye S, Hazzan M, Pottier N, Perrais M, Aubert S, Cauffiez C (2013) Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS ONE 8:e58014. https://doi.org/10.1371/journal.pone.0058014
Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M (2012) microRNA are central players in anti- and profibrotic gene regulation during liver fibrosis. Front Physiol 3:49. https://doi.org/10.3389/fphys.2012.00049
Wang T, Feng Y, Sun H, Zhang L, Hao L, Shi C, Wang J, Li R, Ran X, Su Y, Zou Z (2012) miR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am J Pathol 181:1911–1920. https://doi.org/10.1016/j.ajpath.2012.08.022
Zhu HY, Li C, Bai WD, Su LL, Liu JQ, Li Y, Shi JH, Cai WX, Bai XZ, Jia YH, Zhao B, Wu X, Li J, Hu DH (2014) MicroRNA-21 regulates hTERT via PTEN in hypertrophic scar fibroblasts. PLoS ONE 9:e97114. https://doi.org/10.1371/journal.pone.0097114
Yang X, Wang J, Guo SL, Fan KJ, Li J, Wang YL, Teng Y, Yang X (2011) miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int J Biol Sci 7:685–690. https://doi.org/10.7150/ijbs.7.685
Liu Y, Yang D, Xiao Z, Zhang M (2012) miRNA expression profiles in keloid tissue and corresponding normal skin tissue. Aesthetic Plast Surg 36:193–201. https://doi.org/10.1007/s00266-011-9773-1
Oliveira GV, Hawkins HK, Chinkes D, Burke A, Tavares AL, Ramos-e-Silva M, Albrecht TB, Kitten GT, Herndon DN (2009) Hypertrophic versus non hypertrophic scars compared by immunohistochemistry and laser confocal microscopy: type I and III collagens. Int Wound J 6:445–452. https://doi.org/10.1111/j.1742-481X.2009.00638.x
Rao KB, Malathi N, Narashiman S, Rajan ST (2014) Evaluation of myofibroblasts by expression of alpha smooth muscle actin: a marker in fibrosis, dysplasia and carcinoma. J Clin Diagn Res 8:ZC14-17. https://doi.org/10.7860/JCDR/2014/7820.4231
Shinde AV, Humeres C, Frangogiannis NG (2017) The role of alpha-smooth muscle actin in fibroblast-mediated matrix contraction and remodeling. Biochim Biophys Acta Mol Basis Dis 1863:298–309. https://doi.org/10.1016/j.bbadis.2016.11.006
Arakawa M, Hatamochi A, Takeda K, Ueki H (1990) Increased collagen synthesis accompanying elevated m-RNA levels in cultured Werner’s syndrome fibroblasts. J Invest Dermatol 94:187–190
Mu S, Kang B, Zeng W, Sun Y, Yang F (2016) MicroRNA-143-3p inhibits hyperplastic scar formation by targeting connective tissue growth factor CTGF/CCN2 via the Akt/mTOR pathway. Mol Cell Biochem 416:99–108. https://doi.org/10.1007/s11010-016-2699-9
Zhang D, Liu E, Kang J, Yang X, Liu H (2017) MiR-3613-3p affects cell proliferation and cell cycle in hepatocellular carcinoma. Oncotarget 8:93014–93028. https://doi.org/10.18632/oncotarget.21745
Yan W, Yang W, Liu Z, Wu G (2018) Characterization of microRNA expression in primary human colon adenocarcinoma cells (SW480) and their lymph node metastatic derivatives (SW620). Onco Targets Ther 11:4701–4709. https://doi.org/10.2147/OTT.S169233
Fricke A, Cimniak AFV, Ullrich PV, Becherer C, Bickert C, Pfeifer D, Heinz J, Stark GB, Bannasch H, Braig D, Eisenhardt SU (2018) Whole blood miRNA expression analysis reveals miR-3613-3p as a potential biomarker for dedifferentiated liposarcoma. Cancer Biomark 22:199–207. https://doi.org/10.3233/CBM-170496
Singh AK, Rooge SB, Varshney A, Vasudevan M, Bhardwaj A, Venugopal SK, Trehanpati N, Kumar M, Geffers R, Kumar V, Sarin SK (2018) Global microRNA expression profiling in the liver biopsies of hepatitis B virus-infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology 67:1695–1709. https://doi.org/10.1002/hep.29690
Jiao H, Zheng Z, Shuai X, Wu L, Chen J, Luo Y, Zhao Y, Wang H, Huang Q (2019) MicroRNA expression profiles from HEK293 cells expressing H5N1 avian influenza virus non-structural protein 1. Innate Immun 25:110–117. https://doi.org/10.1177/1753425919826342
Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, Smalheiser NR (2015) Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS ONE 10:e0139233. https://doi.org/10.1371/journal.pone.0139233
Boratyn E, Nowak I, Horwacik I, Durbas M, Mistarz A, Kukla M, Kaczowka P, Lastowska M, Jura J, Rokita H (2016) Monocyte chemoattractant protein-induced protein 1 overexpression modulates transcriptome, including microRNA, in human neuroblastoma cells. J Cell Biochem 117:694–707. https://doi.org/10.1002/jcb.25354
Patil KS, Basak I, Dalen I, Hoedt E, Lange J, Lunde KA, Liu Y, Tysnes OB, Forsgren L, Aarsland D, Neubert TA, Larsen JP, Alves G, Moller SG (2019) Combinatory microRNA serum signatures as classifiers of Parkinson’s disease. Parkinsonism Relat Disord. https://doi.org/10.1016/j.parkreldis.2019.04.010
Thomson BJ (2001) Viruses and apoptosis. Int J Exp Pathol 82:65–76
Upton JW, Chan FK (2014) Staying alive: cell death in antiviral immunity. Mol Cell 54:273–280. https://doi.org/10.1016/j.molcel.2014.01.027
Ambegaokar SS, Roy B, Jackson GR (2010) Neurodegenerative models in Drosophila: polyglutamine disorders, Parkinson disease, and amyotrophic lateral sclerosis. Neurobiol Dis 40:29–39. https://doi.org/10.1016/j.nbd.2010.05.026
Doty RL (2017) Olfactory dysfunction in neurodegenerative diseases: is there a common pathological substrate? Lancet Neurol 16:478–488. https://doi.org/10.1016/S1474-4422(17)30123-0
Zhang D, Jiang P, Xu Q, Zhang X (2011) Arginine and glutamate-rich 1 (ARGLU1) interacts with mediator subunit 1 (MED1) and is required for estrogen receptor-mediated gene transcription and breast cancer cell growth. J Biol Chem 286:17746–17754. https://doi.org/10.1074/jbc.M110.206029
Magomedova L, Tiefenbach J, Zilberman E, Le Billan F, Voisin V, Saikali M, Boivin V, Robitaille M, Gueroussov S, Irimia M, Ray D, Patel R, Xu C, Jeyasuria P, Bader GD, Hughes TR, Morris QD, Scott MS, Krause H, Angers S, Blencowe BJ, Cummins CL (2019) ARGLU1 is a transcriptional coactivator and splicing regulator important for stress hormone signaling and development. Nucleic Acids Res 47:2856–2870. https://doi.org/10.1093/nar/gkz010
Funding
This study was supported by the internal funding source (departmental funding) in Xingtai People’s Hospital.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Research involving human participants and/or animals
All the protocols were approved by the Ethics Committee of Xingtai People’s Hospital. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
All participants in this study were informed and gave a written consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, L., Han, W., Chen, Y. et al. MiR-3613-3p inhibits hypertrophic scar formation by down-regulating arginine and glutamate-rich 1. Mol Cell Biochem 476, 1025–1036 (2021). https://doi.org/10.1007/s11010-020-03968-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03968-4