Abstract
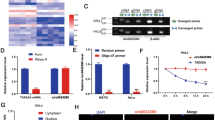
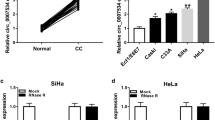
The aim of this study was to investigate the role of circular RNA-9119 (circ9119) in cervical cancer (CC) and the microRNA-126-3p (miR-126)-based molecular mechanism underlying CC. circ9119 and MDM4 were initially overexpressed, and miR-126 expression was found to be reduced in CC cells and tissues. A series of mimics, inhibitors, overexpressing plasmids or siRNAs were introduced into CC cells to alter the circ9119, miR-126, and MDM4 expressions. Cell-based experiments showed that silencing of circ9119 or the upregulation of miR-126 resulted in suppressed proliferation, accompanied by the induced apoptosis of CC cells. The dual-luciferase reporter assay highlighted that circ9119 functioned as an miR-126 ceRNA to increase MDM4 expression. In vivo experiments further confirmed the suppressed tumor growth caused by circ9119 silencing. Our findings demonstrated that circ9119 acts as an oncogene in CC. Our study provides evidence for targeting circ9119 for the treatment of CC.







Similar content being viewed by others
References
Chaturvedi AK (2010) Beyond cervical cancer: burden of other HPV-related cancers among men and women. J Adolesc Health 46:S20–S26
Pimple S, Mishra G, Shastri S (2016) Global strategies for cervical cancer prevention. Curr Opin Obstet Gynecol 28:4–10
Wentzensen N, Schiffman M (2018) Accelerating cervical cancer control and prevention. Lancet Public Health 3:e6–e7
Stewart E, McEvoy J, Wang H et al (2018) Identification of therapeutic targets in rhabdomyosarcoma through integrated genomic, epigenomic, and proteomic analyses. Cancer Cell 34(411–426):e19
Sankaranarayanan R, Ferlay J (2006) Worldwide burden of gynaecological cancer: the size of the problem. Best Pract Res Clin Obstet Gynaecol 20:207–225
Liu X, Zhang L, Liu Y et al (2018) Circ-8073 regulates CEP55 by sponging miR-449a to promote caprine endometrial epithelial cells proliferation via the PI3K/AKT/mTOR pathway. Biochim Biophys Acta (BBA)-Mol Cell Res 1865:1130–1147
He Q, Tian L, Jiang H et al (2017) Identification of laryngeal cancer prognostic biomarkers using an inflammatory gene-related, competitive endogenous RNA network. Oncotarget 8:9525
Huang J, Zhou Q, Li Y (2019) Circular RNAs in gynecological disease: promising biomarkers and diagnostic targets. Biosci Rep. https://doi.org/10.1042/BSR20181641
Chaichian S, Shafabakhsh R, Mirhashemi SM et al (2019) Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol 235(2):718–724
Tang Q, Chen Z, Zhao L (2019) Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (Albany NY) 11(22):9982–9999
Chen RX, Liu HL, Yang LL et al (2019) Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci 23:8771–8778
Ma H, Tian T, Liu X et al (2019) Upregulated circ_0005576 facilitates cervical cancer progression via the miR-153/KIF20A axis. Biomed Pharmacother 118:109311
Fujii T, Shimada K, Nakai T et al (2018) MicroRNAs in smoking-related carcinogenesis: biomarkers, functions, and therapy. J Clin Med 7:98
Rong D, Sun H, Li Z et al (2017) An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8:73271
Xu J, Wang H, Wang H et al (2019) The inhibition of miR-126 in cell migration and invasion of cervical cancer through regulating ZEB1. Hereditas 156:11
Pecorelli S (2009) Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynecol Obstet 105:103–104
Friedman RC, Farh KK-H, Burge CB et al (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105
Yu Q, Liu S-L, Wang H et al (2013) miR-126 Suppresses the proliferation of cervical cancer cells and alters cell sensitivity to the chemotherapeutic drug bleomycin. Asian Pac J Cancer Prev 14:6569–6572
Huang T, Chu TJO (2014) Repression of miR-126 and upregulation of adrenomedullin in the stromal endothelium by cancer-stromal cross talks confers angiogenesis of cervical cancer. Oncogene 33:3636
Li X-M, Wang A-M, Zhang J et al (2011) Down-regulation of miR-126 expression in colorectal cancer and its clinical significance. Med Oncol 28:1054–1057
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384
Qin L, Lin J, Xie X (2019) CircRNA-9119 suppresses poly I: C induced inflammation in Leydig and Sertoli cells via TLR3 and RIG-I signal pathways. Mol Med 25:28
Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321
Lu J, Getz G, Miska EA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Fish JE, Santoro MM, Morton SU et al (2008) miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15:272–284
Lagos-Quintana M, Rauhut R, Yalcin A et al (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12:735–739
Landgraf P, Rusu M, Sheridan R et al (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414
Feng R, Chen X, Yu Y et al (2010) miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett 298:50–63
Zhou Y, Feng X, Liu YL et al (2013) Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS ONE 8:e81203
Du C, Lv Z, Cao L et al (2014) MiR-126-3p suppresses tumor metastasis and angiogenesis of hepatocellular carcinoma by targeting LRP6 and PIK3R2. J Transl Med 12:259
Hamada S, Satoh K, Fujibuchi W et al (2012) MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res 10:3–10
Otsubo T, Akiyama Y, Hashimoto Y et al (2011) MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS ONE 6:e16617
Yang Z, Wang R, Zhang T et al (2015) MicroRNA-126 regulates migration and invasion of gastric cancer by targeting CADM1. Int J Clin Exp Pathol 8:8869–8880
Fang M, Simeonova I, Bardot B et al (2014) Mdm4 loss in mice expressing a p53 hypomorph alters tumor spectrum without improving survival. Oncogene 33:1336
Hoffman Y, Pilpel Y, Oren M (2014) microRNAs and Alu elements in the p53–Mdm2–Mdm4 regulatory network. J Mol Cell Biol 6:192–197
Swetzig WM, Wang J, Das GM (2016) Estrogen receptor alpha (ERα/ESR1) mediates the p53-independent overexpression of MDM4/MDMX and MDM2 in human breast cancer. Oncotarget 7:16049
Kriegmair MC, Balk M, Wirtz R et al (2016) Expression of the p53 inhibitors MDM2 and MDM4 as outcome predictor in muscle-invasive bladder cancer. Anticancer Res 36:5205–5213
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
The procedures conducted in this study were approved by the Ethics Committee of Linyi Central Hospital. Animal experiments were performed under the approval of the Animal Ethics Committee of Linyi Central Hospital.
Informed consent
All subjects signed informed consent forms before their enrollment in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tian, Y., Xu, Z. & Fu, J. CircularRNA-9119 promotes the proliferation of cervical cancer cells by sponging miR-126/MDM4. Mol Cell Biochem 470, 53–62 (2020). https://doi.org/10.1007/s11010-020-03745-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03745-3