Abstract
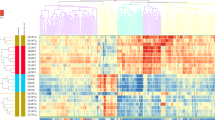
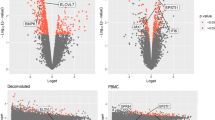
To identify PBMC-expressed genes significant for RA, and to ascertain their upstream regulatory factors, as well as downstream functional effects relevant to RA pathogenesis. We performed peripheral blood mononuclear cells (PBMCs) transcriptome-wide mRNA expression profiling in a case–control discovery sample. Differentially expressed genes (DEGs) were identified and validated in PBMCs in independent samples. We also generated genome-wide SNP genotyping data, and collected miRNA expression data and DNA methylation data from PBMCs of the discovery sample. Pearson correlation analyses were conducted to identify miRNAs/DNA methylations influencing DEG expression. Association analyses were conducted to identify expression-regulating SNPs. The key DEG, SAMD9, which was reported to function as a tumor suppressor gene, was assessed for its effects on T cell proliferation, apoptosis, and inflammatory cytokine expression. A total of 181 DEGs (Fold Change ≥ 2.0, Bonferroni adjusted p ≤ 0.05) were discovered in PBMCs. Four DEGs (SAMD9, CKLF, PARP9, and GUSB), upregulated with RA, were validated independently in PBMCs. Specifically, SAMD9 mRNA expression level was significantly upregulated in PHA-activated Jurkat T cells in vitro, and correlated with 8 miRNAs and associated with 22 SNPs in PBMCs in vivo. Knockdown of SAMD9 could transiently promote Jurkat T cell proliferation within 48 h and significantly induce TNF-α and IL-8 expression in T cells. SAMD9 expression is (epi-) genetically regulated, and significantly upregulated in PBMCs in RA patients and in activated T cells in vitro. SAMD9 might serve as a T cell activation marker but act as an anti-inflammatory factor.



Similar content being viewed by others
References
MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, Silman AJ (2000) Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum 43:30–37
Tedeschi SK, Bermas B, Costenbader KH (2013) Sexual disparities in the incidence and course of SLE and RA. Clin Immunol 149:211–218
Biswas S, Manikandan J, Pushparaj PN (2011) Decoding the differential biomarkers of Rheumatoid arthritis and Osteoarthritis: a functional genomics paradigm to design disease specific therapeutics. Bioinformation 6:153–157
van der Linden MP, Feitsma AL, le Cessie S, Kern M, Olsson LM, Raychaudhuri S, Begovich AB, Chang M, Catanese JJ, Kurreeman FA, van Nies J, van der Heijde DM, Gregersen PK, Huizinga TW, Toes RE, van der Helm-Van Mil AH (2009) Association of a single-nucleotide polymorphism in CD40 with the rate of joint destruction in rheumatoid arthritis. Arthritis Rheum 60:2242–2247
(2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678. https://doi.org/10.1038/nature05911
Morgan AW, Robinson JI, Conaghan PG, Martin SG, Hensor EM, Morgan MD, Steiner L, Erlich HA, Gooi HC, Barton A, Worthington J, Emery P (2010) Evaluation of the rheumatoid arthritis susceptibility loci HLA-DRB1, PTPN22, OLIG3/TNFAIP3, STAT4 and TRAF1/C5 in an inception cohort. Arthritis Res Ther 12:30
Maseda D, Bonami RH, Crofford LJ (2014) Regulation of B lymphocytes and plasma cells by innate immune mechanisms and stromal cells in rheumatoid arthritis. Expert Rev Clin Immunol 10:747–762. https://doi.org/10.1586/1744666x.2014.907744
Chowdhury K, Kumar U, Das S, Chaudhuri J, Kumar P, Kanjilal M, Ghosh P, Sircar G, Basyal RK, Kanga U, Bandyopadhaya S, Mitra DK (2018) Synovial IL-9 facilitates neutrophil survival, function and differentiation of Th17 cells in rheumatoid arthritis. Arthritis Res Ther 20:017–1505
Kurowska W, Kuca-Warnawin E, Radzikowska A, Jakubaszek M, Maslinska M, Kwiatkowska B, Maslinski W (2018) Monocyte-related biomarkers of rheumatoid arthritis development in undifferentiated arthritis patients - a pilot study. Reumatologia 56:10–16. https://doi.org/10.5114/reum.2018.74742
Ishigaki K, Shoda H, Kochi Y, Yasui T, Kadono Y, Tanaka S, Fujio K, Yamamoto K (2015) Quantitative and qualitative characterization of expanded CD4 + T cell clones in rheumatoid arthritis patients. Sci Rep 5:12937. https://doi.org/10.1038/srep12937
Bui VL, Brahn E (2018) Cytokine targeting in rheumatoid arthritis. Clin Immunol. https://doi.org/10.1016/j.clim.2018.04.001
Paludan SR (2000) Synergistic action of pro-inflammatory agents: cellular and molecular aspects. J Leukoc Biol 67:18–25
Bjornsson HT, Fallin MD, Feinberg AP (2004) An integrated epigenetic and genetic approach to common human disease. Trends Genet 20:350–358
Grabiec AM, Reedquist KA (2013) The ascent of acetylation in the epigenetics of rheumatoid arthritis. Nat Rev Rheumatol 9(5):311–318. https://doi.org/10.1038/nrrheum.2013.17
Ehrlich M, Lacey M (2013) DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics 5:553–568. https://doi.org/10.2217/epi.13.43
Xiao C, Rajewsky K (2009) MicroRNA control in the immune system: basic principles. Cell 136:26–36
Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N (2010) MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4 + T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol 184:6773–6781
Klein K, Gay S (2015) Epigenetics in rheumatoid arthritis. Curr Opin Rheumatol 27:76–82. https://doi.org/10.1097/bor.0000000000000128
He P, Xia W, Wang L, Wu J, Guo YF, Zeng KQ, Wang MJ, Bing PF, Xie FF, Lu X, Zhang YH, Lei SF, Deng FY (2018) Identification of expression quantitative trait loci (eQTLs) in human peripheral blood mononuclear cells (PBMCs) and shared with liver and brain. J Cell Biochem 119:1659–1669
Goeman JJ, Solari A (2014) Multiple hypothesis testing in genomics. Stat Med 33:1946–1978
Zhao H, Nyholt DR, Yang Y, Wang J, Yang Y (2017) Improving the detection of pathways in genome-wide association studies by combined effects of SNPs from linkage disequilibrium blocks. Sci Rep 7:3512. https://doi.org/10.1038/s41598-017-03826-2
Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43:5
Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44:839–847
Wang L, Zhu J, Deng FY, Wu LF, Mo XB, Zhu XW, Xia W, Xie FF, He P, Bing PF, Qiu YH, Lin X, Lu X, Zhang L, Yi NJ, Zhang YH, Lei SF (2018) Correlation analyses revealed global microRNA-mRNA expression associations in human peripheral blood mononuclear cells. Mol Genet Genomics 293:95–105
Xie FF, Deng FY, Wu LF, Mo XB, Zhu H, Wu J, Guo YF, Zeng KQ, Wang MJ, Zhu XW, Xia W, Wang L, He P, Bing PF, Lu X, Zhang YH, Lei SF (2018) Multiple correlation analyses revealed complex relationship between DNA methylation and mRNA expression in human peripheral blood mononuclear cells. Funct Integr Genomics 18:1–10. https://doi.org/10.1007/s10142-017-0568-6
Jouy F, Muller SA, Wagner J, Otto W, von Bergen M, Tomm JM (2015) Integration of conventional quantitative and phospho-proteomics reveals new elements in activated Jurkat T-cell receptor pathway maintenance. Proteomics 15:25–33. https://doi.org/10.1002/pmic.201400119
Lin YP, Su CC, Huang JY, Lin HC, Cheng YJ, Liu MF, Yang BC (2009) Aberrant integrin activation induces p38 MAPK phosphorylation resulting in suppressed Fas-mediated apoptosis in T cells: implications for rheumatoid arthritis. Mol Immunol 46:3328–3335. https://doi.org/10.1016/j.molimm.2009.07.021
Lee EY, Seo M, Juhnn YS, Kim JY, Hong YJ, Lee YJ, Lee EB, Song YW (2011) Potential role and mechanism of IFN-gamma inducible protein-10 on receptor activator of nuclear factor kappa-B ligand (RANKL) expression in rheumatoid arthritis. Arthritis Res Ther 13:R104. https://doi.org/10.1186/ar3385
Li T, Zhong J, Chen Y, Qiu X, Zhang T, Ma D, Han W (2006) Expression of chemokine-like factor 1 is upregulated during T lymphocyte activation. Life Sci 79:519–524
Bramwell KK, Ma Y, Weis JH, Chen X, Zachary JF, Teuscher C, Weis JJ (2014) Lysosomal beta-glucuronidase regulates Lyme and rheumatoid arthritis severity. J Clin Invest 124:311–320
Zhu H, Wu LF, Mo XB, Lu X, Tang H, Zhu XW, Xia W, Guo YF, Wang MJ, Zeng KQ, Wu J, Qiu YH, Lin X, Zhang YH, Liu YZ, Yi NJ, Deng FY, Lei SF (2019) Rheumatoid arthritis-associated DNA methylation sites in peripheral blood mononuclear cells. Ann Rheum Dis 78:36–42. https://doi.org/10.1136/annrheumdis-2018-213970
Lemos de Matos A, Liu J, McFadden G, Esteves PJ (2013) Evolution and divergence of the mammalian SAMD9/SAMD9L gene family. BMC Evol Biol 13:1471–2148
Tanaka M, Shimbo T, Kikuchi Y, Matsuda M, Kaneda Y (2010) Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int J Cancer 126:1982–1991. https://doi.org/10.1002/ijc.24965
Topaz O, Indelman M, Chefetz I, Geiger D, Metzker A, Altschuler Y, Choder M, Bercovich D, Uitto J, Bergman R, Richard G, Sprecher E (2006) A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am J Hum Genet 79:759–764
Bodolay E, Koch AE, Kim J, Szegedi G, Szekanecz Z (2002) Angiogenesis and chemokines in rheumatoid arthritis and other systemic inflammatory rheumatic diseases. J Cell Mol Med 6:357–376
Szekanecz Z, Vegvari A, Szabo Z, Koch AE (2010) Chemokines and chemokine receptors in arthritis. Front Biosci 2:153–167
Mateen S, Zafar A, Moin S, Khan AQ, Zubair S (2016) Understanding the role of cytokines in the pathogenesis of rheumatoid arthritis. Clin Chim Acta 455:161–171
Juarranz MG, Santiago B, Torroba M, Gutierrez-Canas I, Palao G, Galindo M, Abad C, Martinez C, Leceta J, Pablos JL, Gomariz RP (2004) Vasoactive intestinal peptide modulates proinflammatory mediator synthesis in osteoarthritic and rheumatoid synovial cells. Rheumatology 43:416–422. https://doi.org/10.1093/rheumatology/keh061
Chefetz I, Ben Amitai D, Browning S, Skorecki K, Adir N, Thomas MG, Kogleck L, Topaz O, Indelman M, Uitto J, Richard G, Bradman N, Sprecher E (2008) Normophosphatemic familial tumoral calcinosis is caused by deleterious mutations in SAMD9, encoding a TNF-alpha responsive protein. J Invest Dermatol 128:1423–1429
Li CF, MacDonald JR, Wei RY, Ray J, Lau K, Kandel C, Koffman R, Bell S, Scherer SW, Alman BA (2007) Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics 8:1471–2164
Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavare S, Deloukas P, Dermitzakis ET (2007) Population genomics of human gene expression. Nat Genet 39:1217–1224
van Baarsen LG, Wijbrandts CA, Rustenburg F, Cantaert T, van der Pouw Kraan TC, Baeten DL, Dijkmans BA, Tak PP, Verweij CL (2010) Regulation of IFN response gene activity during infliximab treatment in rheumatoid arthritis is associated with clinical response to treatment. Arthritis Res Ther 12:22
Sanda C, Weitzel P, Tsukahara T, Schaley J, Edenberg HJ, Stephens MA, McClintick JN, Blatt LM, Li L, Brodsky L, Taylor MW (2006) Differential gene induction by type I and type II interferons and their combination. J Interferon Cytokine Res 26:462–472
Hershkovitz D, Gross Y, Nahum S, Yehezkel S, Sarig O, Uitto J, Sprecher E (2011) Functional characterization of SAMD9, a protein deficient in normophosphatemic familial tumoral calcinosis. J Invest Dermatol 131:662–669
Mekhedov SL, Makarova KS, Koonin EV (2017) The complex domain architecture of SAMD9 family proteins, predicted STAND-like NTPases, suggests new links to inflammation and apoptosis. Biol Direct 12:017–0185
Acknowledgements
The study was supported by Natural Science Foundation of China (81373010, 81473046, 81541068, 31271336, 81502868, 31401079 and 81401343), the Natural Science Foundation of Jiangsu Province (BK20150346), the Startup Fund from Soochow University (Q413900112, Q413900712) and a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Institutional Research Ethic Board at the Soochow University (ethics approval number: 2012-146). All the participants signed informed consent form.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, P., Wu, LF., Bing, PF. et al. SAMD9 is a (epi-) genetically regulated anti-inflammatory factor activated in RA patients. Mol Cell Biochem 456, 135–144 (2019). https://doi.org/10.1007/s11010-019-03499-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-019-03499-7