Abstract
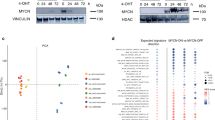
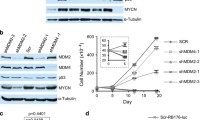
c-Myc is a highly pleiotropic transcription factor known to control cell cycle progression, apoptosis, and cellular transformation. Normally, ectopic expression of c-Myc is associated with promoting cell proliferation or triggering cell death via activating p53. However, it is not clear how the levels of c-Myc lead to different cellular responses. Here, we generated a series of stable RPE cell clones expressing c-Myc at different levels, and found that consistent low level of c-Myc induced cellular senescence by activating AP4 in post-confluent RPE cells, while the cells underwent cell death at high level of c-Myc. In addition, high level of c-Myc could override the effect of AP4 on cellular senescence. Further knockdown of AP4 abrogated senescence-like phenotype in cells expressing low level of c-Myc, and accelerated cell death in cells with medium level of c-Myc, indicating that AP4 was required for cellular senescence induced by low level of c-Myc.






Similar content being viewed by others
References
Lowe SW, Cepero E, Evan G (2004) Intrinsic tumor suppression. Nature 432:307–315
Collado M, Serrano M (2010) Senescence in tumors: evidence from mice and humans. Nat Rev Cancer 10:51–57
Campaner S, Doni M, Verrecchia A, Fagà G, Bianchi L, Amati B (2010) Myc, Cdk2 and cellular senescence: old players, new game. Cell Cycle 9:3655–3661
Nesbit CE, Tersak JM, Prochownik EV (1999) MYC oncogenes and human neoplastic disease. Oncogene 18:3004–3016
Pelengaris S, Khan M, Evan G (2002) c-MYC: more than just a matter of life and death. Nat Rev Cancer 2:764–776
Dang CV, Lee LA (1995) c-Myc function in neoplasia. Springer. ISBN 9783540606321
Dang CV (2013) MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 10.1101/cshperspect.a014217
Galaktionov K, Chen X, Beach D (1996) Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382:511–517
Hoffman B, Amanullah A, Shafarenko M, Liebermann DA (2002) The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene 21:3414–3421
Delgado MD, León J (2010) Myc roles in hematopoiesis and leukemia. Genes Cancer 1:605–616
Peschle C, Mavilio F, Sposi NM, Giampaolo A, Caré A, Bottero L, Bruno M, Mastroberardino G, Gastaldi R, Testa MG (1984) Translocation and rearrangement of c-myc into immunoglobulin a heavy chain locus in primary cells from acute lymphocytic leukemia. Proc Natl Acad Sci USA 81:5514–5518
Escot C, Theillet C, Lidereau R, Spyratos F, Champeme MH, Gest J, Callahan R (1986) Genetic alteration of the c-myc proto-oncogene (MYC) in human primary breast carcinomas. Proc Natl Acad Sci USA 83:4834–4838
Schlotter CM, Vogt U, Bosse U, Mersch B, Wassmann K (2003) C-myc, not HER-2/neu, can predict recurrence and mortality of patients with node-negative breast cancer. Breast Cancer Res 5:R30–R36
Felsher DW, Bishop JM (1999) Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 4:199–207
Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW (2002) Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297:102–104
Marinkovic D, Marinkovic T, Mahr B, Hess J, Wirth T (2004) Reversible lymphomagenesis in conditionally c-MYC expressing mice. Int J Cancer 110:336–342
Pelengaris S, Littlewood T, Khan M, Elia G, Evan G (1999) Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell 3:565–577
Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW (2007) Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci USA 104:13028–13033
Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI (2008) Modelling Myc inhibition as a cancer therapy. Nature 455:679–683
Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, Frank S, Amati B, Dalla-Favera R, Monnat RJ Jr (2003) Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev 17:1569–1574
Robinson K, Asawachaicharn N, Galloway DA, Grandori C (2009) c-Myc accelerates S-phase and requires WRN to avoid replication stress. PLoS ONE 4:e5951
Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjö S, Murga M, Fernandez-Capetillo O, Barbacid M, Larsson LG, Amati B (2010) Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol 12:54–59
Reimann M, Lee S, Loddenkemper C, Dörr JR, Tabor V, Aichele P, Stein H, Dörken B, Jenuwein T, Schmitt CA (2010) Tumor stroma-derived TGF-beta limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17:262–272
van Riggelen J, Müller J, Otto T, Beuger V, Yetil A, Choi PS, Kosan C, Möröy T, Felsher DW, Eilers M (2010) The interaction between Myc and Miz1 is required to antagonize TGF-beta-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev 24:1281–1294
Mermod N, Williams TJ, Tjian R (1988) Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature 332:557–561
Hu YF, Lüscher B, Admon A, Mermod N, Tjian R (1990) Transcription factor AP-4 contains multiple dimerization domains that regulate dimer specificity. Genes Dev 4:1741–1752
Badinga L, Song S, Simmen RC, Simmen FA (1998) A distal regulatory region of the insulin-like growth factor binding protein-2 (IGFBP-2) gene interacts with the basic helix–loop–helix transcription factor, AP4. Endocrine 8:281–289
Tsujimoto K, Ono T, Sato M, Nishida T, Oguma T, Tadakuma T (2005) Regulation of the expression of caspase-9 by the transcription factor activator protein-4 in glucocorticoid-induced apoptosis. J Biol Chem 280:27638–27644
Jung P, Menssen A, Mayr D, Hermeking HP (2008) AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci USA 105:15046–15051
Jackstadt R, Röh S, Neumann J, Jung P, Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A, Hermeking H (2013) AP4 is a mediator of epithelial–mesenchymal transition and metastasis in colorectal cancer. J Exp Med 210:1331–1350
Friess H, Ding J, Kleeff J, Fenkell L, Rosinski JA, Guweidhi A, Reidhaar-Olson JF, Korc M, Hammer J, Büchler MW (2003) Microarray-based identification of differentially expressed growth- and metastasis-associated genes in pancreatic cancer. Cell Mol Life Sci 60:1180–1199
Xinghua L, Bo Z, Yan G, Lei W, Changyao W, Qi L, Lin Y, Kaixiong T, Guobin W, Jianying C (2012) The overexpression of AP4 as a prognostic indicator for gastric carcinoma. Med Oncol 29:871–877
Huang Q, Raya A, DeJesus P, Chao SH, Quon KC, Caldwell JS, Chanda SK, Izpisua-Belmonte JC, Schultz PG (2004) Identification of p53 regulators by genome-wide functional analysis. Proc Natl Acad Sci USA 101:3456–3461
Wang Y, Wong MM, Zhang X, Chiu SK (2015) Ectopic AP4 expression induces cellular senescence via activation of p53 in long-term confluent retinal pigment epithelial cells. Exp Cell Res 339:135–146
Chen S, Chiu SK (2015) AP4 activates cell migration and EMT mediated by p53 in MDA-MB-231 breast carcinoma cells. Mol Cell Biochem 407:57–68
Shi L, Jackstadt R, Siemens H, Li H, Kirchner T, Hermeking H (2014) p53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial–mesenchymal transition and metastasis in colorectal cancer. Cancer Res 74:532–542
Binder S, Stanzel BV, Krebs I, Glittenberg C (2007) Transplantation of the RPE in AMD. Prog Retin Eye Res 26:516–554
Proulx S, Landreville S, Guérin SL, Salesse C (2004) Integrin alpha5 expression by the ARPE-19 cell line: comparison with primary RPE cultures and effect of growth medium on the alpha5 gene promoter strength. Exp Eye Res 79:157–165
Pfeffer BA, Philp NJ (2014) Cell culture of retinal pigment epithelium: special Issue. Exp Eye Res 126:1–4
Ricci MS, Jin Z, Dews M, Yu D, Thomas-Tikhonenko A, Dicker DT, El-Deiry WS (2004) Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol 24:8541–8555
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O et al (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92:9363–9367
Leontieva OV, Demidenko ZN, Blagosklonny MV (2014) Contact inhibition and high cell density deactivate the mammalian target of rapamycin pathway, thus suppressing the senescence program. Proc Natl Acad Sci USA 111:8832–8837
Liao W, Ning G (2006) Knockdown of apolipoprotein B, an atherogenic apolipoprotein, in HepG2 cells by lentivirus-mediated siRNA. Biochem Biophys Res Commun 344:478–483
Danielian PS, White R, Hoare SA, Fawell SE, Parker MG (1993) Identification of residues in the estrogen receptor that confer differential sensitivity to estrogen and hydroxytamoxifen. Mol Endocrinol 7:232–240
Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W (2013) Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Mol Cell 51:46–56
Hoffman B, Liebermann DA (2008) Apoptotic signaling by c-MYC. Oncogene 27:6462–6472
Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC (1992) Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119–128
Hydbring P, Bahram F, Su Y, Tronnersjo S, Hogstrand K, von der Lehr N, Sharifi HR, Lilischkis R, Hein N, Wu S, Vervoorts J, Henrikssona M, Grandien A, Lüscher B, Larsson L (2010) Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci USA 107:58–63
Hydbring P, Larsson LG (2010) Tipping the balance: Cdk2 enables Myc to suppress senescence. Cancer Res 70:6687–6691
Liu X, Zhang B, Guo Y, Liang Q, Wu C, Wu L, Tao K, Wang G, Chen J (2012) Down-regulation of AP-4 inhibits proliferation, induces cell cycle arrest and promotes apoptosis in human gastric cancer cells. PLoS ONE 7:e37096
Hu X, Guo W, Chen S, Xu Y, Li P, Wang H, Chu H, Li J, Du Y, Chen X, Zhang G, Zhao G (2016) Silencing of AP-4 inhibits proliferation, induces cell cycle arrest and promotes apoptosis in human lung cancer cells. Oncol Lett 11:3735–3742
Philipp A, Schneider A, Väsrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M (1994) Repression of cyclin D1: a novel function of MYC. Mol Cell Biol 14:4032–4043
Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O’Connell BC, Mateyak MK, Tam W, Kohlhuber F, Dang CV, Sedivy JM, Eick D, Vogelstein B, Kinzler KW (2000) Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA 97:2229–2234
Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM (2002) c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell 9:1031–1044
Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R (2007) Non-transcriptional control of DNA replication by c-Myc. Nature 448:445–451
Menssen A, Epanchintsev A, Lodygin D, Rezaei N, Jung P, Verdoodt B, Diebold J, Hermeking H (2007) c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1: identification of mechanisms underlying c-MYC-induced DNA damage and chromosomal instability. Cell Cycle 6:339–352
Felsher DW, Bishop JM (1999) Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA 96:3940–3944
Jackstadt R, Hermeking H (2014) AP4 is required for mitogen- and c-MYC-induced cell cycle progression. OncoTarget 5:7316–7327
Reisman D, Elkind NB, Roy B, Beamon J, Rotter V (1993) c-Myc trans-activates the p53 promoter through a required downstream CACGTG motif. Cell Growth Differ 4:57–65
Acknowledgements
This work was funded by the Seed Grant from City University of Hong Kong (Project Number 7004028), the National Natural Science Foundation of China (No. 81703753), the Research Fund of Zhejiang Chinese Medicine University (No. 2017ZR05), and the NSERC Discovery Grant NSERC (RGPIN-2015-04144).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Rights and permissions
About this article
Cite this article
Wang, Y., Cheng, X., Samma, M.K. et al. Differential cellular responses by oncogenic levels of c-Myc expression in long-term confluent retinal pigment epithelial cells. Mol Cell Biochem 443, 193–204 (2018). https://doi.org/10.1007/s11010-017-3224-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-017-3224-5