Abstract
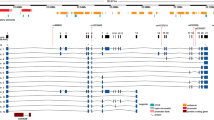
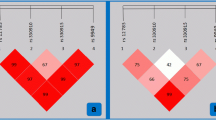
Plasma level of cyclophilin A is a promising marker of vascular disease in patients with type 2 diabetes. Genetic variants in the peptidylprolyl isomerase A gene, encoding human cyclophilin may alter protein synthesis thus affecting its activity, function, and circulating plasma levels. We examined the effect of single-nucleotide polymorphisms (SNPs) within the PPIA gene on plasma levels of cyclophilin A and coupled this with status of vascular disease in patients with and without type 2 diabetes in 212 South Indian subjects. The regulatory region of PPIA gene was sequenced for SNPs. The association of SNPs with known blood markers of type 2 diabetes and coronary artery disease such as HbA1c, low- and high-density lipoproteins, triglycerides, fasting and postprandial blood sugar levels, and cyclophilin A were probed. We identified three SNPs namely, rs6850: A > G; (AG/−) c.*227_*228delAG and (−/T) c.*318_*319insT. Welchs two-sample t test indicated an association of SNP rs6850: A > G, located at the 5′ UTR region with increased plasma levels of cyclophilin A in patients with coronary artery disease and with coronary artery disease associated with diabetes. The presence of rs6850: A > G variant was significantly associated with coronary artery disease irrespective of whether the patients had diabetes or not. In silico analysis of the sequence using different tools and matrix libraries did not predict any significant differential binding sites for rs6850: A > G, c.*227_*228delAG and c.*318_*319insT. Our results indicate that the SNP rs6850: A > G is associated with increased risk for elevated plasma levels of cyclophilin A and coronary artery disease in patients with and without type 2 diabetes.


Similar content being viewed by others
Abbreviations
- PPIA:
-
Peptidyl-prolyl isomerase A
- CAD:
-
Coronary artery disease
- DM:
-
Diabetes mellitus
- DM + CAD:
-
Diabetes mellitus + coronary artery disease
- UTR:
-
Untranslated region
- EDTA:
-
Ethylene diamine tetra acetic acid
- ELISA:
-
Enzyme-linked immunosorbent assay
- FBS:
-
Fasting blood sugar
- HbA1c:
-
Glycated hemoglobin
- HDL:
-
High-density lipoprotein
- LDL:
-
Low-density lipoprotein
- Tris HCl:
-
Tris hydrochloric acid
- NaCl:
-
Sodium chloride
- SDS:
-
Sodium dodecyl sulfate
- TRANSFAC:
-
Transcription Factor database
- ANOVA:
-
Analysis of variance
- CI:
-
Confidence intervals
- OR:
-
Odds ratios
- CPBP:
-
Core promoter binding protein
- LEF-1:
-
Lymphoid enhancer-binding factor-1
- VSMCs:
-
Vascular smooth muscle cells
- GWAS:
-
Genome-wide association study
- TFBS:
-
Transcription factor binding sites
References
Ramachandran S, Venugopal A, Kutty VR et al (2014) Plasma level of cyclophilin A is increased in patients with type 2 diabetes mellitus and suggests presence of vascular disease. Cardiovasc Diabetol 13:38
Bergsma DJ, Eder C, Gross M et al (1991) The cyclophilin multigene family of peptidyl-prolyl isomerases characterization of three separate human isoforms. J Biol Chem 266:23204–23214
Matouschek A, Rospert S, Schmid K et al (1995) Cyclophilin catalyzes protein folding in yeast mitochondria. Proc Natl Acad Sci USA 92:6319–6323
Nigro P, Pompilio G, Capogrossi MC (2013) Cyclophilin A: a key player for human disease. Cell Death Dis 4:e888
Pflügl G, Kallen J, Schirmer T et al (1993) X-ray structure of a decameric cyclophilin-cyclosporin crystal complex. Nature 361:91–94
Haendler B, Hofer E (1990) Characterization of the human cyclophilin gene and of related processed pseudogenes. Eur J Biochem 190:477–482
Ghatak S, Muthukumaran RB, Nachimuthu SK (2013) Simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. J Biomol Tech 24:224–231
An P, Wang LH, Hutcheson-Dilks H et al (2007) Regulatory polymorphisms in the cyclophilin A gene, PPIA accelerate progression to AIDS. PLoS Pathog 3:e88
Rebhan M, Chalifa Caspi V, Prilusky J et al (1998) GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 14:656–664
Safran M, Chalifa Caspi V, Shmueli O, Olender T et al (2003) Human gene-centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW21, and HORDE. Nucl Acids Res 31:142–146
Surendran S, Girijamma A, Nair R et al (2014) Forkhead box C2 promoter variant c.-512C.T is associated with increased susceptibility to chronic venous diseases. PLoS One 9:e90682
Matys V, Kel-Margoulis OV, Fricke E et al (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucl Acids Res 34:D108–D110
Berger MF, Philippakis AA, Qureshi AM et al (2006) Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24:1429–1435
Messeguer X, Escudero R, Farré D et al (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18:333–334
Farre D, Roset R, Huerta M et al (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucl Acids Res 31:3651–3653
Chang TH, Huang HY, Hsu JB et al (2013) An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinform 14:2105–2114
Agarwal V, Bell GW, Nam J et al (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna 2012
Palacín M, Rodríguez I, García-Castro M et al (2008) A search for cyclophilin-A gene (PPIA) variation and its contribution to the risk of atherosclerosis and myocardial infarction. Int J Immunogenet 35:159–164
Ramachandran S, Venugopal A, Sathisha K et al (2012) Proteomic profiling of high glucose primed monocytes identifies cyclophilin A as a potential secretory marker of inflammation in type 2 diabetes. Proteomics 12:2808–2821
Ley K, Miller YI, Hedrick CC (2011) Monocyte and Macrophage Dynamics during Atherogenesis. Arterioscler Thromb Vasc Biol 31:1506–1516
Billinch A, Winkier G, Aschauer H et al (1997) Presence of cyclophilin A in synovial fluids of patients with rheumatoid arthritis. J Exp Med 185:975–980
Satoh K, Nigro P, Berk BC (2010) Oxidative stress and vascular smooth muscle cell growth: a mechanistic linkage by Cyclophilin A. Antioxid Redox Signal 12:675–682
Soe NN, Sowden M, Baskaran P et al (2014) Acetylation of cyclophilin A is required for its secretion and vascular cell activation. Cardiovasc Res 101:444–453
Ramachandran S, Kartha CC (2012) Cyclophilin-A: a potential screening marker for vascular disease in type-2 diabetes. Can J Physiol Pharmacol 90:1005–1015
Jin ZG, Berk BC (2004) Role of secreted oxidative stress induced factors (SOXFs) in the pathogenesis of atherosclerosis. Arch Mal Coeur Vaiss 97:1256–1259
Schunkert H, Konig IR, Thompson J et al (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43:333–338
Preuss M, Konig JR, Thompson JR et al (2010) Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: a genome-wide association meta-analysis involving more than 22,000 cases and 60,000 controls. Circ Cardiovasc Genet 3:475–483
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14000 cases of seven common diseases and 3000 shared controls. Nature 447:661–678
Erdmann J, Grosshennig A, Braund PS et al (2009) New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet 41:280–282
Gudbjartsson DF, Bjornsdottir US, Halap E et al (2009) Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet 41:342–347
Tregouet DA, Konig IR, Erdmann J et al (2009) Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet 41:283–285
Coronary Artery Disease (C4D) Genetics Consortium (2011) A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 43:339–344
Slavin D, Sapin V, Lopez-Diaz F et al (2014) The Krüppel-like core promoter binding protein gene is primarily expressed in placenta during mouse development. Oncotarget 5:649–658
Albano F, Zagaria A, Anelli L et al (2013) Lymphoid enhancer binding factor-1 (LEF1) expression as a prognostic factor in adult acute promyelocytic leukemia. Oncotarget 5:649–658
Sonis ST, Keefe DM (2013) Pathobiology of cancer regimen related toxicities. Springer, New York. doi:10.1007/978-1-4614-5438
Bleiber G, May M, Martinez R et al (2005) Use of a combined ex vivo/in vivo population approach for screening of human genes involved in the human immunodeficiency virus type 1 life cycle for variants influencing disease progression. J Virol 79:12674–12680
Rezzani R, Favero G, Stacchiotti A et al (2013) Endothelial and vascular smooth muscle cell dysfunction mediated by cyclophilin A and the atheroprotective effects of melatonin. Life Sci 92:875–882
Satoh K, Nigro P, Matoba T et al (2009) Cyclophilin A enhances vascular oxidative stress and the development of angiotensin II-induced aortic aneurysms. Nat Med 15:649–656
Seizer P, Schonberger T, Schott M et al (2010) EMMPRIN and its ligand cyclophilin A regulate MT1-MMP, MMP-9 and M-CSF during foam cell formation. Atherosclerosis 209:51–57
Coppinger JA, Cagney G, Toomey S et al (2004) Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood 103:2096–2104
Elvers M, Herrmann A, Seizer P et al (2012) Intracellular cyclophilin A is an important Ca(2þ) regulator in platelets and critically involved in arterial thrombus formation. Blood 120:1317–1326
Acknowledgments
This work was supported by Indian Council of Medical Research, Ministry of Health, Government of India (No 5/4/1-4/2013/NCD-II). Financial support from Director, Rajiv Gandhi Centre for Biotechnology is also acknowledged.
Authors contributions
VA drafted the manuscript and carried out all molecular biology experiments; RK designed, analyzed, and interpreted statistical data. VKA and RG performed bioinformatics analysis and interpreted data. DG and SS were involved in acquisition of data and drafting the manuscript. KRS, NSP, and AM contributed to design of the study and acquisition of data. CCK and SR were equally involved in conceiving the study, its design, interpretation of results, and critically reviewed and revised the manuscript before submission. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no competing interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vinitha, A., Kutty, V.R., Vivekanand, A. et al. PPIA rs6850: A > G single-nucleotide polymorphism is associated with raised plasma cyclophilin A levels in patients with coronary artery disease. Mol Cell Biochem 412, 259–268 (2016). https://doi.org/10.1007/s11010-015-2632-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2632-7