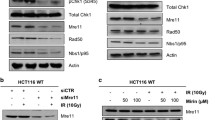
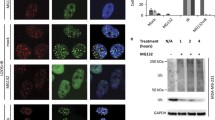
Abstract
ING proteins are tumor suppressors involved in the regulation of gene transcription, cell cycle arrest, apoptosis, and senescence. Here, we show that ING1b expression is upregulated by several DNA-damaging agents, in a p53-independent manner. ING1b stimulates DNA repair of a variety of DNA lesions requiring activation of multiple DNA repair pathways. Moreover, Ing1 −/− cells showed impaired genomic DNA repair after H2O2 and neocarzinostatin treatment and this defect was reverted by overexpression of ING1b. Two tumor-derived ING1 mutants failed to promote DNA repair highlighting the physiological importance of the integrity of the PHD domain for ING1b DNA repair activity and suggesting a role in the prevention of tumor progression. Ing−/− cells showed higher basal levels of γ-H2AX and, upon DNA damage, γ-H2AX increase was greater and with faster kinetics compared to wild-type cells. Chromatin relaxation by Trichostatin A led to an exacerbated damage signal in both types of cells, but this effect was dependent on Ing1 status, and more pronounced in wild-type cells. Our results suggest that ING1 acts at early stages of the DNA damage response activating a variety of repair mechanisms and that this function of ING1 is targeted in tumors.






Similar content being viewed by others
Abbreviations
- ING1:
-
Inhibitor of growth 1
- DDR:
-
DNA damage response
- NCS:
-
Neocarzinostatin
- UDS:
-
Unscheduled DNA synthesis
- PHD:
-
Plant homeodomain
References
Coles AH, Jones SN (2009) The ING gene family in the regulation of cell growth and tumorigenesis. J Cell Physiol 218:45–57
Li X, Kikuchi K, Takano Y (2011) ING genes work as tumor suppressor genes in the carcinogenesis of head and neck squamous cell carcinoma. J Oncol. doi:10.1155/2011/963614
Soliman MA, Riabowol K (2007) After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci 32:509–519
Nouman GS, Anderson JJ, Lunec J, Angus B (2003) The role of the tumour suppressor p33 ING1b in human neoplasia. J Clin Pathol 56:491–496
Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R (2008) The new tumor suppressor genes ING: genomic structure and status in cancer. Int J Cancer 123:1483–1490
Champagne KS, Kutateladze TG (2009) Structural insight into histone recognition by the ING PHD fingers. Curr Drug Targets 10:432–441
Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J (2006) ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 21:51–64
Ludwig S, Klitzsch A, Baniahmad A (2011) The ING tumor suppressors in cellular senescence and chromatin. Cell Biosci 1: 25
Abad M, Moreno A, Palacios A, Narita M, Blanco F, Moreno-Bueno G, Narita M and Palmero I (2011) The tumor suppressor ING1 contributes to epigenetic control of cellular senescence. Aging Cell 10: 158–171
Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK (2004) The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature 428:328–332
Gomez-Cabello D, Callejas S, Benguria A, Moreno A, Alonso J and Palmero I (2010) Regulation of the microRNA processor DGCR8 by the tumor suppressor ING1. Cancer Res 70: 1866–1874
Nozell S, Laver T, Moseley D, Nowoslawski L, De Vos M, Atkinson GP, Harrison K, Nabors LB, Benveniste EN (2008) The ING4 tumor suppressor attenuates NF-kappaB activity at the promoters of target genes. Mol Cell Biol 28:6632–6645
Xin H, Yoon HG, Singh PB, Wong J, Qin J (2004) Components of a pathway maintaining histone modification and heterochromatin protein 1 binding at the pericentric heterochromatin in Mammalian cells. J Biol Chem 279:9539–9546
Abad M, Menendez C, Fuchtbauer A, Serrano M, Fuchtbauer EM, Palmero I (2007) Ing1 mediates p53 accumulation and chromatin modification in response to oncogenic stress. J Biol Chem 282:31060–31067
Huertas D, Sendra R, Munoz P (2009) Chromatin dynamics coupled to DNA repair. Epigenetics 4:31–42
Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, Riabowol K (2001) UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci 114:3455–3462
Wong RP, Lin H, Khosravi S, Piche B, Jafarnejad SM, Chen DW, Li G (2011) Tumour suppressor ING1b maintains genomic stability upon replication stress. Nucleic Acids Res 39: 3632–3642
Cheung KJ, Jr., Mitchell D, Lin P, Li G (2001) The tumor suppressor candidate p33(ING1) mediates repair of UV-damaged DNA. Cancer Res 61: 4974–4977
Kuo WH, Wang Y, Wong RP, Campos EI, Li G (2007) The ING1b tumor suppressor facilitates nucleotide excision repair by promoting chromatin accessibility to XPA. Exp Cell Res 313:1628–1638
Campos EI, Martinka M, Mitchell DL, Dai DL, Li G (2004) Mutations of the ING1 tumor suppressor gene detected in human melanoma abrogate nucleotide excision repair. Int J Oncol 25:73–80
Garate M, Campos EI, Bush JA, Xiao H, Li G (2007) Phosphorylation of the tumor suppressor p33(ING1b) at Ser-126 influences its protein stability and proliferation of melanoma cells. FASEB J 21:3705–3716
Goeman F, Thormeyer D, Abad M, Serrano M, Schmidt O, Palmero I, Baniahmad A (2005) Growth inhibition by the tumor suppressor p33ING1 in immortalized and primary cells: involvement of two silencing domains and effect of Ras. Mol Cell Biol 25:422–431
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 162:156–159
Palmero I, Murga M, Zubiaga A, Serrano M (2002) Activation of ARF by oncogenic stress in mouse fibroblasts is independent of E2F1 and E2F2. Oncogene 21:2939–2947
Gonzalez L, Freije JM, Cal S, Lopez-Otin C, Serrano M, Palmero I (2006) A functional link between the tumour suppressors ARF and p33ING1. Oncogene 25:5173–5179
Cheung KJ Jr., Bush JA, Jia W, Li G (2000) Expression of the novel tumour suppressor p33(ING1)is independent of p53. Br J Cancer 83: 1468–1472
Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K (2000) Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res 60:3143–3146
Chen L, Matsubara N, Yoshino T, Nagasaka T, Hoshizima N, Shirakawa Y, Naomoto Y, Isozaki H, Riabowol K, Tanaka N (2001) Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res 61:4345–4349
Rogakou EP, Boon C, Redon C, Bonner WM (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146:905–916
Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of the genome. DNA Repair (Amst) 3:959–967
Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE (2007) Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol 178:209–218
Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O (2007) Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol 178:1101–1108
Skowyra D, Zeremski M, Neznanov N, Li M, Choi Y, Uesugi M, Hauser CA, Gu W, Gudkov AV, Qin J (2001) Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J Biol Chem 276:8734–8739
Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol Cell Biol 22:835–848
Vieyra D, Loewith R, Scott M, Bonnefin P, Boisvert FM, Cheema P, Pastyryeva S, Meijer M, Johnston RN, Bazett-Jones DP, McMahon S, Cole MD, Young D, Riabowol K (2002) Human ING1 proteins differentially regulate histone acetylation. J Biol Chem 277:29832–29839
Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV (1998) The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391:295–298
Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y, Appella E, Yokota J, Harris CC (2003) p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res 63:2373–2378
Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC (2001) DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci USA 98:9671–9676
Nagashima M, Shiseki M, Pedeux RM, Okamura S, Kitahama-Shiseki M, Miura K, Yokota J, Harris CC (2003) A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene 22:343–350
Pedeux R, Sengupta S, Shen JC, Demidov ON, Saito S, Onogi H, Kumamoto K, Wincovitch S, Garfield SH, McMenamin M, Nagashima M, Grossman SR, Appella E, Harris CC (2005) ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol Cell Biol 25:6639–6648
Doyon Y, Selleck W, Lane WS, Tan S, Cote J (2004) Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 24:1884–1896
Kichina JV, Zeremski M, Aris L, Gurova KV, Walker E, Franks R, Nikitin AY, Kiyokawa H, Gudkov AV (2006) Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 25:857–866
Tsang FC, Po LS, Leung KM, Lau A, Siu WY, Poon RY (2003) ING1b decreases cell proliferation through p53-dependent and -independent mechanisms. FEBS Lett 553:277–285
Kataoka H, Bonnefin P, Vieyra D, Feng X, Hara Y, Miura Y, Joh T, Nakabayashi H, Vaziri H, Harris CC, Riabowol K (2003) ING1 represses transcription by direct DNA binding and through effects on p53. Cancer Res 63:5785–5792
Tamannai M, Farhangi S, Truss M, Sinn B, Wurm R, Bose P, Henze G, Riabowol K, von Deimling A, Tallen G (2010) The inhibitor of growth 1 (ING1) is involved in trichostatin A-induced apoptosis and caspase 3 signaling in p53-deficient glioblastoma cells. Oncol Res 18: 469–480
Hara Y, Zheng Z, Evans SC, Malatjalian D, Riddell DC, Guernsey DL, Wang LD, Riabowol K, Casson AG (2003) ING1 and p53 tumor suppressor gene alterations in adenocarcinomas of the esophagogastric junction. Cancer Lett 192:109–116
Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R (2007) Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 6:470–480
Hatayama T, Yukioka M (1982) Action of neocarzinostatin on cell nuclei: release of specific chromatin. Biochem Biophys Res Commun 104:889–896
Kappen LS, Ellenberger TE, Goldberg IH (1987) Mechanism and base specificity of DNA breakage in intact cells by neocarzinostatin. Biochemistry 26:384–390
Shi X, Gozani O (2005) The fellowships of the INGs. J Cell Biochem 96:1127–1136
Pena PV, Hom RA, Hung T, Lin H, Kuo AJ, Wong RP, Subach OM, Champagne KS, Zhao R, Verkhusha VV, Li G, Gozani O, Kutateladze TG (2008) Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. J Mol Biol 380:303–312
Gibbons RJ, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, Berry SA, Dahl N, Fryer A, Keppler K, Kurosawa K, Levin ML, Masuno M, Neri G, Pierpont ME, Slaney SF, Higgs DR (1997) Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet 17:146–148
Saugier-Veber P, Drouot N, Wolf LM, Kuhn JM, Frebourg T, Lefebvre H (2001) Identification of a novel mutation in the autoimmune regulator (AIRE-1) gene in a French family with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. Eur J Endocrinol 144:347–351
Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, Gozani OP (2005) A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem 280:28701–28710
Stark M, Puig-Butille JA, Walker G, Badenas C, Malvehy J, Hayward N, Puig S (2006) Mutation of the tumour suppressor p33ING1b is rare in melanoma. Br J Dermatol 155:94–99
Acknowledgments
This study was supported by research grants from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Agencia Nacional de Promoción Científica y Tecnológica (ANPCYT), and Universidad de Buenos Aires to EC and Spanish Ministry of Science and Innovation to IP (SAF2009-09031). We thank Dr. Luciana E. Giono for help with the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
11010_2013_1601_MOESM1_ESM.tif
ESM 1 ING1b improves DNA repair in response to UV. HeLa cells were transfected with a vector encoding wild-type ING1b (ING wt) or mutants ING1bC215S (ING 215), ING1bK270 N (ING 270) along with pBabePuro. Twenty-four hours after transfection regular medium was substituted for arginine-free medium containing 1 % FBS and cells were incubated for additional 60 h in presence of puromycin. Resistant cells were treated with 8 mJ/cm2 UV and incubated with 10 mCi [3H] thymidine for 10 h. Cell lysates were tested for genomic DNA repair by UDS assay. Bars represent the mean ± S.E. of three experiments performed in triplicate. Student t test was used to compare samples (*P < 0.05). (TIFF 1645 kb)
Rights and permissions
About this article
Cite this article
Ceruti, J.M., Ogara, M.F., Menéndez, C. et al. Inhibitor of growth 1 (ING1) acts at early steps of multiple DNA repair pathways. Mol Cell Biochem 378, 117–126 (2013). https://doi.org/10.1007/s11010-013-1601-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-013-1601-2