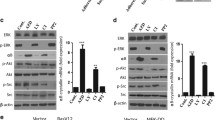
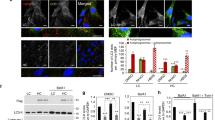
Abstract
The extracellular matrix (ECM) plays a key role in cell–cell communication and signaling, and the signals it propagates are important for tissue remodeling and survival. However, signals from disease-altered ECM may lead to anoikis—apoptotic cell death triggered by loss of ECM contacts. Previously, we found that an altered fibronectin matrix triggers anoikis in human primary ligament cells via a pathway that requires p53 transcriptional downregulation. Here we show that this p53 reduction is suppressed by transfecting cells with Mdm2 antisense oligonucleotides or small interfering RNA. Similar results were found in cells treated to prevent p53 and Mdm2 interactions. When p53 was overexpressed in cells lacking Mdm2 and p53, p53 levels were unaffected by anoikis conditions. However, cells cotransfected with p53 and wild type Mdm2, but not a mutant Mdm2, exhibited decreased p53 levels in response to anoikis conditions. Thus, cells under anoikis conditions undergo p53 degradation that is mediated by Mdm2.




Similar content being viewed by others
Notes
The abbreviations used are: ECM, extracellular matrix; cAFn and AFn, alternatively spliced V region (V+) and containing intact (H+) or a mutated, nonfunctional (H−) high-affinity heparin binding domain; TBST, 25 mM Tris, 150 mM NaCl, 0.05% Tween-20.
References
Homandberg GA, Wen C, Hui F (1998) Cartilage damaging activities of fibronectin fragments derived from cartilage and synovial fluid. Osteoarthr Cartil 6:231–244
Huynh QN, Wang S, Tafolla E, Gansky SA, Kapila S, Armitage GC, Kapila YL (2002) Specific fibronectin fragments as markers of periodontal disease status. J Periodontol 73:1101–1110
Talonpoika J, Heino J, Larjava H, Hakkinen L, Paunio K (1989) Gingival crevicular fluid fibronectin degradation in periodontal health and disease. Scand J Dent Res 97:415–421
Talonpoika J, Paunio K, Soderling E (1993) Molecular forms and concentration of fibronectin and fibrin in human gingival crevicular fluid before and after periodontal treatment. Scand J Dent Res 101:375–381
Xie DL, Meyers R, Homandberg GA (1992) Fibronectin fragments in osteoarthritic synovial fluid. J Rheumatol 19:1448–1452
Homandberg GA, Meyers R, Williams JM (1993) Intraarticular injection of fibronectin fragments causes severe depletion of cartilage proteoglycans in vivo. J Rheumatol 20:1378–1382
Homandberg GA, Meyers R, Xie DL (1992) Fibronectin fragments cause chondrolysis of bovine articular cartilage slices in culture. J Biol Chem 267:3597–3604
Kapila YL, Kapila S, Johnson PW (1996) Fibronectin and fibronectin fragments modulate the expression of proteinases and proteinase inhibitors in human periodontal ligament cells. Matrix Biol 15:251–261
Kapila YL, Lancero H, Johnson PW (1998) The response of periodontal ligament cells to fibronectin. J Periodontol 69:1008–1019
Kapila YL, Niu J, Johnson PW (1997) The high affinity heparin-binding domain and the V region of fibronectin mediate invasion of human oral squamous cell carcinoma cells in vitro. J Biol Chem 272:18932–18938
Kapila YL, Wang S, Dazin P, Tafolla E, Mass MJ (2002) The heparin-binding domain and V region of fibronectin regulate apoptosis by suppression of p53 and c-myc in human primary cells. J Biol Chem 277:8482–8491
Kapila YL, Wang S, Johnson PW (1999) Mutations in the heparin binding domain of fibronectin in cooperation with the V region induce decreases in pp125(FAK) levels plus proteoglycan-mediated apoptosis via caspases. J Biol Chem 274:30906–30913
Tafolla E, Wang S, Wong B, Leong J, Kapila YL (2005) JNK1 and JNK2 oppositely regulate p53 in signaling linked to apoptosis triggered by an altered fibronectin matrix: JNK links FAK and p53. J Biol Chem 280:19992–19999
Xie D, Homandberg GA (1993) Fibronectin fragments bind to and penetrate cartilage tissue resulting in proteinase expression and cartilage damage. Biochim Biophys Acta 1182:189–196
Dai R, Iwama A, Wang S, Kapila YL (2005) Disease-associated fibronectin matrix fragments trigger anoikis of human primary ligament cells: p53 and c-myc are suppressed. Apoptosis 10:503–512
Forsyth CB, Pulai J, Loeser RF (2002) Fibronectin fragments and blocking antibodies to alpha2beta1 and alpha5beta1 integrins stimulate mitogen-activated protein kinase signaling and increase collagenase 3 (matrix metalloproteinase 13) production by human articular chondrocytes. Arthritis Rheum 46:2368–2376
Gemba T, Valbracht J, Alsalameh S, Lotz M (2002) Focal adhesion kinase and mitogen-activated protein kinases are involved in chondrocyte activation by the 29-kDa amino-terminal fibronectin fragment. J Biol Chem 277:907–911
Homandberg GA, Costa V, Wen C (2002) Fibronectin fragments active in chondrocytic chondrolysis can be chemically cross-linked to the alpha5 integrin receptor subunit. Osteoarthr Cartil 10:938–949
Yasuda T, Kakinuma T, Julovi SM, Yoshida M, Hiramitsu T, Akiyoshi M, Nakamura T (2004) COOH-terminal heparin-binding fibronectin fragment induces nitric oxide production in rheumatoid cartilage through CD44. Rheumatology 43:1116–1120
Brooks CL, Gu W (2006) p53 ubiquitination: Mdm2 and beyond. Mol Cell 21:307–315
Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408:307–310
Yang Y, Li CC, Weissman AM (2004) Regulating the p53 system through ubiquitination. Oncogene 23:2096–2106
Michael D, Oren M (2003) The p53–Mdm2 module and the ubiquitin system. Semin Cancer Biol 13:49–58
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2:169–178
Mani A, Gelmann EP (2005) The ubiquitin–proteasome pathway and its role in cancer. J Clin Oncol 23:4776–4789
Ghosh A, Joo NE, Chen TC, Kapila YL (2010) Proapoptotic fibronectin fragment induces the degradation of ubiquitinated p53 via proteasomes in periodontal ligament cells. J Periodontal Res. doi:10.1111/j.1600-0765.2009.01261.x
Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP (1998) Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol 18:3735–3743
McDonnell TJ, Montes de Oca Luna R, Cho S, Amelse LL, Chavez-Reyes A, Lozano G (1999) Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. J Pathol 188:322–328
Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B (1990) Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912–915
Chen J, Wu X, Lin J, Levine AJ (1996) mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol 16:2445–2452
Zhang Z, Li M, Wang H, Agrawal S, Zhang R (2003) Antisense therapy targeting MDM2 oncogene in prostate cancer: Effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc Natl Acad Sci USA 100:11636–11641
Akhtar S, Kole R, Juliano RL (1991) Stability of antisense DNA oligodeoxynucleotide analogs in cellular extracts and sera. Life Sci 49:1793–1801
Klein C, Vassilev LT (2004) Targeting the p53–MDM2 interaction to treat cancer. Br J Cancer 91:1415–1419
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848
Kumar SK, Hager E, Pettit C, Gurulingappa H, Davidson NE, Khan SR (2003) Design, synthesis, and evaluation of novel boronic-chalcone derivatives as antitumor agents. J Med Chem 46:2813–2815
Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387:299–303
Honda R, Tanaka H, Yasuda H (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420:25–27
Duncan SJ, Gruschow S, Williams DH, McNicholas C, Purewal R, Hajek M, Gerlitz M, Martin S, Wrigley SK, Moore M (2001) Isolation and structure elucidation of Chlorofusin, a novel p53–MDM2 antagonist from a Fusarium sp. J Am Chem Soc 123:554–560
Zhao J, Wang M, Chen J, Luo A, Wang X, Wu M, Yin D, Liu Z (2002) The initial evaluation of non-peptidic small-molecule HDM2 inhibitors based on p53-HDM2 complex structure. Cancer Lett 183:69–77
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke K, Koeppen H, Dixit VM (2004) The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86–92
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S (2003) Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779–791
Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387:296–299
Asher G, Tsvetkov P, Kahana C, Shaul Y (2005) A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev 19:316–321
Harris SL, Levine AJ (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24:2899–2908
Momand J, Zambetti GP, Olson DC, George D, Levine AJ (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237–1245
Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B (1992) Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80–83
Girnita L, Girnita A, Larsson O (2003) Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc Natl Acad Sci USA 100:8247–8252
Kimura SH, Nojima H (2002) Cyclin G1 associates with MDM2 and regulates accumulation and degradation of p53 protein. Genes Cells 7:869–880
Nag A, Bagchi S, Raychaudhuri P (2004) Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res 64:8152–8155
Song MS, Song SJ, Kim SY, Oh HJ, Lim DS (2008) The tumour suppressor RASSF1A promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex. EMBO J 27:1863–1874
Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C (2009) Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35:316–326
Chen D, Kon N, Li M, Zhang W, Qin J, Gu W (2005) ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121:1071–1083
Asher G, Lotem J, Sachs L, Kahana C, Shaul Y (2002) Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc Natl Acad Sci USA 99:13125–13130
Acknowledgments
We thank Dr. Gerard P. Zambetti (St. Jude Children’s Research Hospital, Memphis, TN) for p53-null fibroblasts, Dr. Gigi Lozano (University of Texas M.D. Anderson Cancer Center, Houston, TX) for p53/Mdm2-null fibroblasts, Dr. Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD) for human wildtype p53 and human Mdm2 pCMV expression plasmids, Dr. Arnold J. Levine (Princeton University, Princeton, NJ) for human Mdm2 pCHDMΔ222-437 expression plasmid, Dr. Paul W. Johnson for mentoring and the fibronectin fragments and Stephen Ordway for editorial assistance. This study was support by an NIH grant RO1 DE013725 (to Y. L. K.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghosh, A., Chen, T.C. & Kapila, Y.L. Anoikis triggers Mdm2-dependent p53 degradation. Mol Cell Biochem 343, 201–209 (2010). https://doi.org/10.1007/s11010-010-0514-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-010-0514-6