Abstract
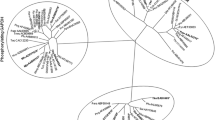
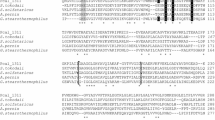
Alignment of the amino acid sequence of some archaeal, bacterial and eukaryotic non-phosphorylating glyceraldehydes-3-phosphate dehydrogenases (GAPNs) and aldehyde dehydrogenases (ALDHs) with the sequence of a putative GAPN present in the genome of the Gram-negative bacterium Neisseria meningitidis strain Z2491 demonstrated the conservation of residues involved in the catalytic activity. The predicted coding sequence of the N. meningitidis gapN gene was cloned in Escherichia coli XL1-blue under the expression of an inducible promoter. The IPTG-induced GAPN was purified ca. 48-fold from E. coli cells using a procedure that sequentially employed conventional ammonium sulfate fractionation as well as anion-exchange and affinity chromatography. The purified recombinant enzyme was thoroughly characterized. The protein is a homotetramer with a 50-kDa subunit, exhibiting absolute specificity for NAD and a broad spectrum of aldehyde substrates. Isoelectric focusing analysis with the purified fraction showed the presence of an acidic polypeptide with an isoelectric point of 6.3. The optimum pH of the purified enzyme was between 9 and 10. Studies on the effect of increasing temperatures on the enzyme activity revealed an optimal value ca. 64 °C. Molecular phylogenetic data suggest that N. meningitidis GAPN has a closer relationship with archaeal GAPNs and glyceraldehyde dehydrogenases than with the typical NADP-specific GAPNs from Gram-positive bacteria and photosynthetic eukaryotes.




Similar content being viewed by others
References
Caugant DA (1998) Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106:505–525
Exley RM, Shaw J, Mowe E et al (2005) Available carbon source influences the resistance of Neisseria meningitidis against complement. J Exp Med 201:637–1645
Forthergill-Gilmore LA, Michels PAM (1993) Evolution of glycolysis. Prog Biophys Biol 95:105–135
Brinkmann H, Cerff R, Salomon M et al (1989) Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinash. Plant Mol Biol 13:81–94
Cerff R (1982) Separation and purification of NAD- and NADP-linked glyceraldehyde-3-phosphate dehydrogenase from higher plants. In: Edekman M, Hallik RB, Chua MH (eds) Methods in chloroplast molecular biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, pp 683–694
Valverde F, Losada M, Serrano A (1997) Functional complementation of an Escherichia coli gap mutant supports an amphibolic role for NAD(P)-dependent glyceraldehyde-3-phosphate dehydrogenase of Synechocystis sp. Strain PCC 6803. J Bacteriol 179:4513–4522
Cerff R (1995) Origin and evolution of phosphorylating and non-phosphorylating glyceraldehyde-3-phosphate dehydrogenases. In: Mathis P (ed) Photosynthesis from light to biosphere, vol 1. Kluwer Academic Publishers, Dordrecht, pp 933–938
Habenicht A, Hellman U, Cerff R (1994) Non-phosphorylating GAPDH of higher plants is a member of the aldehyde dehydrogenase superfamily with no sequence homology to phosphorylating GAPDH. J Mol Biol 237:165–171
Mateos MI, Serrano A (1992) Ocurrence of phosphorylating and non-phosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase in photosynthetic organisms. Plant Sci 84:163–170
Serrano A, Mateos MI, Losada M (1993) ATP-driven transhydrogenation and ionization of water in a reconstituted glyceraldehyde-3-phosphate dehydrogenase (phosphorylating and non-phosphorylating) model system. Biochem Biophys Res Commun 193:1348–1356
Iglesias A, Serrano A, Guerrero MG et al (1987) Purification and properties of NADP-dependent non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from the green alga Chlamydomonas reinhardtii. Biochem Biophys Acta 925:1–10
Boyd DA, Cvitkovitch DG, Hamilton IR (1995) Sequence, expression, and function of the gene for the non-phosphorylating, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase of Streptococcus mutans. J Bacteriol 177:2622–2627
Iddar A, Valverde F, Serrano A et al (2002) Expression, purification and characterization of recombinant non-phosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Clostridium acetobutylicum. Protein Expr Purif 25:519–526
Iddar A, Valverde F, Serrano A et al (2003) Purification of Recombinant non-phosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Streptococcus pyogenes expressed in E. coli. Mol Cell Biochem 247:195–203
Iddar A, Valverde F, Assobhei O et al (2005) Widespread occurrence of non- phosphorylating glyceraldehyde-3-phosphate dehydrogenase among gram-positive bacteria. Int Microbiol 8:251–258
Van der Oost J, Schut G, Kengen SWM et al (1998) The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. Biol Chem 273: 28149–28154
Brunner NA, Brinkmann H, Siebers B et al (1998) NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. J Biol Chem 273:6149–6156
Ahmed H, Ettema TJ, Tjaden B et al (2005) The semi-phosphorylative Entner–Doudoroff pathway in hyperthermophilic archaea: a re-evaluation. Biochem J 390:529–540
Lorentzen E, Hensel R, Knura T et al (2004) Structural basis of allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehydes-3-phosphate dehydrogenase from Thermoproteus tenax. J Mol Biol 341:815–828
Parkhill J, Achtman M, James KD et al (2000) Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502–506
Kellogg DS, Peacock WL, Deacon WE et al (1963) Neisseria gonorrhoeae. Virulence genetically linked to clonal variation. J Bacteriol 85:1274–1279
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Valverde F, Losada M, Serrano A (1999) A Engineering a central metabolic pathway: glycolysis with no net phosphorylating in an Escherichia coli gap complemented with a plant gapN gene. FEBS Lett 449:153–158
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:4668–4673
Hedrick JL, Smith AJ (1968) Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys 162:155–164
Thompson JD, Gibson TJ, Plewniak F et al (1997) The CLUSTALX Windows interface: flexible strategies for multiple sequence aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Strimmer K, von Haeseler A (1996) Quarter puzzling: a quarter maximum likelihood method for reconstructing tree topologies. Mol Biol Evol 13:964–969
Marchal S, Branlant G (1999) Evidence for the chemical activation of essential cys-302 upon cofactor binding to non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans. Biochemistry 38:12950–12958
Marchal S, Cobessi D, Rahuel-Clermont S et al (2001) Chemical mechanism and substrate binding sites of NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. Chem Biol Interact 30:15–28
Marchal S, Rahuel-Clermont S, Branlant G (2000) Role of glutamate- 268 in catalytic mechanism of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans. Biochemistry 39:3327–3335
Liu ZJ, Sun YJ, Rose J et al (1997) The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol 4:317–326
Marchal S, Branlant G (2002) Characterization of the amino acids involved in substrate specificity of nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans. J Biol Chem 277:39235–39242
Harris JI, Waters M (1976) Glyceraldehyde-3-phosphate dehydrogenase. In: Boyer PD (ed) The enzymes, vol 13. Academic Press, New York, pp 1–49
Habenicht A (1997) The non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase: biochemistry, structure, occurrence and evolution. Biol Chem 378:1413–1419
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Jung HJ, Lee SB (2006) Identification and characterization of Thermoplasma acidophilum glyceraldehyde dehydrogenase: a new class of NADP+-specific aldehyde dehydrogenase. Biochem J 397:131–138
Reher M, Schönheit P (2006) Glyceraldehyde dehydrogenases from the thermophilic euryarcheota Picrophilus torridus and Thermoplasma acidophilum, key enzymes of the non-phosphorylative Entner–Doudoroff pathway, constitute a novel enzyme family within the aldehyde dehydrogenase superfamily. FEBS Lett 280:1198–1204
Hafid N, Valverde F, Villalobos E et al (1998) Glyceraldehyde-3-phosphate dehydrogenase from Tetrahymena pyriformis: enzyme purification and characterization of a gapC gene with primitive eukaryotic features. Comp Biochem Physiol 119:493–503
Brunner NA, Lang DA, Wilmanns M et al (2000) Crystallization and preliminary X-ray diffraction analysis of the NAD-dependent non-phosphorylating GAPDH of the hyperthermophilic archaeon Thermoproteus tenax. Acta Cryst 56:89–91
Pohl E, Brunner N, Wilmanns M, Hensel R (2002) The crystal structure of the allosteric non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeum Thermoproteus tenax. J Biol Chem 277:19938–19945
Brown AT, Wittenberger CL (1971) Mechanism for regulating the distribution of glucose carbon between the Embden–Meyerhof and hexosemonophosphate pathways in Streptococcus faecalis. J Bacteriol 106:456–467
Habenicht A, Quesada A, Cerff R (1997) Sequence of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Nicotiana plumbaginifolia and phylogenetic origin of the gene family. Gene 198:237–243
Acknowledgments
The authors thank Prof. M. Losada for encouragement and help. This work is part of several collaborative Research Projects between the CNR (Morocco) and CSIC (Spain). It was supported by AECI (Spain) and collaborative grants of the Andalusian Government—Ministère d’Education et de la Recherche Scientifique of Morocco (Junta de Andalucía, Proyectos de Cooperación al Desarrollo en el Ámbito Universitario de la Agencia Andaluza de Cooperación Internacional A52/02, and A54/04), group PAI CVI-261 and grants from CNRST (Morocco).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fourrat, L., Iddar, A., Valverde, F. et al. Cloning, gene expression and characterization of a novel bacterial NAD-dependent non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Neisseria meningitidis strain Z2491. Mol Cell Biochem 305, 209–219 (2007). https://doi.org/10.1007/s11010-007-9545-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-007-9545-z