Abstract
Recent advances have demonstrated substantial benefits from learning with both generative and discriminative parameters. On the one hand, generative approaches address the estimation of the parameters of the joint distribution—\(\mathrm{P}(y,\mathbf{x})\), which for most network types is very computationally efficient (a notable exception to this are Markov networks) and on the other hand, discriminative approaches address the estimation of the parameters of the posterior distribution—and, are more effective for classification, since they fit \(\mathrm{P}(y|\mathbf{x})\) directly. However, discriminative approaches are less computationally efficient as the normalization factor in the conditional log-likelihood precludes the derivation of closed-form estimation of parameters. This paper introduces a new discriminative parameter learning method for Bayesian network classifiers that combines in an elegant fashion parameters learned using both generative and discriminative methods. The proposed method is discriminative in nature, but uses estimates of generative probabilities to speed-up the optimization process. A second contribution is to propose a simple framework to characterize the parameter learning task for Bayesian network classifiers. We conduct an extensive set of experiments on 72 standard datasets and demonstrate that our proposed discriminative parameterization provides an efficient alternative to other state-of-the-art parameterizations.
Similar content being viewed by others
1 Introduction
Efficient training of Bayesian Network Classifiers has been the topic of much recent research (Buntine 1994; Carvalho et al. 2011; Friedman et al. 1997; Heckerman and Meek 1997; Martinez et al. 2016; Pernkopf and Bilms 2010; Webb et al. 2012; Zaidi et al. 2013). Two paradigms predominate (Jebara 2003). One can optimize the log-likelihood (LL). This is traditionally called generative learning. The goal is to obtain parameters characterizing the joint distribution in the form of local conditional distributions and then estimate class-conditional probabilities using Bayes rule. Alternatively, one can optimize the conditional-log-likelihood (CLL)—known as discriminative learning. The goal is to directly estimate the parameters associated with the class-conditional distribution—\(\mathrm{P}(y|\mathbf{x})\).
Naive Bayes (NB) is a Bayesian network \({ \mathop { \text {BN} } }\) that specifies independence between attributes given the class. Recent work has shown that placing a per-attribute-value-per-class-value weight on probabilities in NB (and learning these weights by optimizing the CLL) leads to an alternative parameterization of vanilla Logistic Regression (LR) (Zaidi et al. 2014). The introduction of these weights (and optimizing them by maximizing CLL) also makes it possible to relax NB’s conditional independence assumption and thus to create a classifier with lower bias (Ng and Jordan 2002; Zaidi et al. 2014). The classifier is low-biased, as weights can remedy inaccuracies introduced by invalid attribute-independence assumptions.
In this paper, we generalize this idea to the general class of \({ \mathop { \text {BN} } }\) classifiers. Like NB, any given \({ \mathop { \text {BN} } }\) structure encodes assumptions about conditional independencies between the attributes and will result in error if they do not hold in the data. Optimizing the log-likelihood in this case will result in suboptimal performance for classification (Friedman et al. 1997; Grossman and Domingos 2004; Su et al. 2008) and one should either optimize directly the CLL by learning the parameters of the class-conditional distribution or by placing weights on the probabilities and learn these weights by optimizing the CLL.
The main contributions of this paper are:
-
1.
We develop a new discriminative parameter learning method for Bayesian network classifiers by combining fast generative parameter (and structure) learning with subsequent fast discriminative parameter estimation (using parameter estimates from the former to precondition search for the parameters of the latter). To achieve this, discriminative parameters are restated as weights rectifying deviations of the discriminative model from the generative one (in terms of the violation of independence between factors present in the generative model).
-
2.
A second contribution of this work is the development of a simple framework to characterize the parameter learning task for Bayesian network classifiers. Building on previous work by Friedman et al. (1997), Greiner et al. (2005), Pernkopf and Wohlmayr (2009), Roos et al. (2005) and Zaidi et al. (2013), this framework allows us to lay out the different techniques in a systematic manner; highlighting similarities, distinctions and equivalences.
Our proposed parameterization is based on a two-step learning process:
-
1.
Generative step: We maximize the LL to obtain parameters for all local conditional distributions in the \({ \mathop { \text {BN} } }\).
-
2.
Discriminative step: We associate a weight with each parameter learned in the generative step and re-parameterize the class-conditional distribution in terms of these weights (and of the fixed generative parameters). We can then discriminatively learn these weights by optimizing the CLL.
In this paper, we show that:
-
The proposed formalization of the parameter learning task for \({ \mathop { \text {BN} } }\) is actually a re-parameterization of the one step (discriminative) learning problem (this will become clear when we introduce the proposed framework) but with faster convergence of the discriminative optimization procedure. In the experimental section, we complement our theoretical framework with an empirical analysis over 72 domains; the results demonstrate the superiority of our approach. In Sect. 5.5, we will discuss our proposed approach from the perspective of pre-conditioning in unconstrained optimization problems.
-
The proposed approach results in a three-level hierarchy of nested parameterizations, where each additional level introduces (or “unties”) exponentially more parameters in order to fit ever smaller violations of independence.
-
Regularization of the discriminative parameters in the proposed discriminative learning approach allows to limit the amount of allowable violation of independence and effectively interpolate between discriminative and generative parameter estimation.
The rest of this paper is organized as follows. In Sect. 2, we present our proposed framework for parameter learning of Bayesian network classifiers. We also give the formulation for class-conditional Bayesian Network models (CCBN) in this section. Two established parameterizations of class-conditional Bayesian networks are given in Sects. 3 and 4, respectively. In Sect. 5, we present our proposed parameterization of CCBN. In Sect. 6, we discuss some related work to this research. Experimental analysis is conducted in Sect. 7. We conclude in Sect. 8 with some pointers to future work.
All the symbols used in this work are listed in Table 1.
2 A simple framework for parameter learning of \({ \mathop { \text {BN} } }\) classifiers
We start by discussing Bayesian network classifiers in the following section.
2.1 Bayesian network classifiers
A \({ \mathop { \text {BN} } }\) \({\mathcal {B}}= \langle {\mathcal {G}},\varTheta \rangle \), is characterized by the structure \({\mathcal {G}}\) (a directed acyclic graph, where each vertex is a variable, \(Z_i\)), and a set of parameters \(\varTheta \), that quantifies the dependencies within the structure. The variables are partitioned into a single target, the class variable \(Y{=}Z_0\) and n covariates \(X_1{=}Z_1, X_2{=}Z_2, \ldots X_n{=}Z_n\), called the attributes. The parameter \(\varTheta \), contains a set of parameters for each vertex in \({\mathcal {G}}\): \(\theta _{z_0|\varPi _0(\mathbf{x})}\) and for \(1\le i\le n\), \(\theta _{z_i|y,\varPi _i(\mathbf{x})}\), where \(\varPi _i(.)\) is a function which given the datum \(\mathbf{x}= \langle x_1, x_1,\ldots ,x_n \rangle \) as its input, returns the values of the attributes that are the parents of node i in structure \({\mathcal {G}}\). For notational simplicity, instead of writing \(\theta _{Z_0 = z_0 | \varPi _0(\mathbf{x})}\) and \(\theta _{Z_i = z_i | y,\varPi _i(\mathbf{x})}\), we write \(\theta _{z_0|\varPi _0(\mathbf{x})}\) and \(\theta _{z_i|y,\varPi _i(\mathbf{x})}\). A \({ \mathop { \text {BN} } }\) \({\mathcal {B}}\) computes the joint probability distribution as: \(\mathrm{P}_{\mathcal {B}}(y,\mathbf{x}) = \theta _{z_0 | \varPi _0(\mathbf{x})}\cdot \prod _{i=1}^{n} \theta _{z_i |y, \varPi _i(\mathbf{x})}\).
For a \({ \mathop { \text {BN} } }\) \({\mathcal {B}}\), we can write:
Now, the corresponding conditional distribution \(\mathrm{P}_{\mathcal {B}}(y|\mathbf{x})\) can be computed with the Bayes rule as:
If the class attribute does not have any parents, we write: \(\theta _{y|\varPi _0(\mathbf{x})} = \theta _y\).
Given a set of data points \(\mathcal {D}= \{ \mathbf{x}^{(1)},\ldots ,\mathbf{x}^{(N)} \}\), the Log-Likelihood (LL) of \({\mathcal {B}}\) is:
Maximizing Eq. 3 to optimize the parameters (\(\theta \)) is the maximum-likelihood estimation of the parameters.
Theorem 1
Within the constraints in Eq. 4, Eq. 3 is maximized when \(\theta _{x_i | \varPi _i(\mathbf{x})}\) corresponds to empirical estimates of probabilities from the data, that is, \(\theta _{y | \varPi _0(\mathbf{x})} = \mathrm{P}_\mathcal {D}(y | \varPi _0(\mathbf{x}))\) and \(\theta _{x_i | \varPi _i(\mathbf{x})} = \mathrm{P}_\mathcal {D}(x_i | \varPi _i(\mathbf{x}))\).
Proof
See “Appendix 1”. \(\square \)
The parameters obtained by maximizing Eq. 3 (and fulfilling the constraints in Eq. 4) are typically known as ‘Generative’ estimates of the probabilities.
2.2 Class-conditional BN (CCBN) models
Instead of following a two-step process for classification with \({ \mathop { \text {BN} } }\), where step 1 involves maximizing \(\mathrm{P}(y,\mathbf{x})\) and the second step is application of Bayes rule to obtain \(\mathrm{P}(y|\mathbf{x})\), one can directly optimize for \(\mathrm{P}(y|\mathbf{x})\) by maximizing the Conditional Log-Likelihood (CLL). Optimizing CLL is generally considered a more effective objective function (for classification) since it directly optimizes the mapping from features to class labels. The CLL can be defined as:
which is equal to:
The only difference between Eqs. 3 and 5 is the presence of the normalization factor in the latter, that is: \(\log \sum _{y'}^{|\mathcal {Y}|} \mathrm{P}_{{\mathcal {B}}}(y',\mathbf{x}^{(j)})\). Due to this normalization, the values of \(\theta \) maximizing Eq. 5 are not the same as those that maximize Eq. 3. We provide two intuitions as to why maximizing the CLL should provide a better model of the conditional distribution:
-
1.
It allows the parameters to be set in such a way as to reduce the effect of the conditional attribute independence assumption that is present in the BN structure and that might be violated in data.
-
2.
We have \(\text {LL}({\mathcal {B}})=\text {CLL}({\mathcal {B}})+\text {LL}({\mathcal {B}}\backslash y)\). If optimizing \(\text {LL}({\mathcal {B}})\), most of the attention will be given to \(\text {LL}({\mathcal {B}}\backslash y)\)—because \(\text {CLL}({\mathcal {B}})\ll \text {LL}({\mathcal {B}}\backslash y)\)—which will often lead to poor estimates for classification.
Note, that if the structure is correct, maximizing both LL and CLL should lead to the same results (Rubinstein and Hastie 1997). There is unfortunately no closed-form solution for \(\theta \) such that the CLL would be maximized; we thus have to resort to numerical optimization methods over the space of parameters.
Like any Bayesian network model, a class-conditional \({ \mathop { \text {BN} } }\) model is composed of a graphical structure and of parameters (\({\varvec{\theta }}\)) quantifying the dependencies in the structure. For any \({ \mathop { \text {BN} } }\) \({\mathcal {B}}\), the corresponding CC\({ \mathop { \text {BN} } }\) will be based on graph \({\mathcal {B}^{*}}\) (where \({\mathcal {B}^{*}}\) is a sub-graph of \({\mathcal {B}}\)) whose parameters are optimized by maximizing the CLL. We present below a slightly rephrased definition from Roos et al. (2005):
Definition 1
A class-conditional Bayesian network model \({\mathcal {M}^{{\mathcal {B}}^*}}\) is the set of conditional distributions based on the network \({\mathcal {B}}^*\) equipped with any strictly positive parameter set \({\varvec{\theta }}^{{\mathcal {B}}^*}\); that is the set of all functions from \((X_1,X_2,\ldots ,X_n)\) to a distribution on Y takes the form of Eq. 2.
This means that the nodes in \({\mathcal {B}}^*\) are nodes comprising only the Markov blanket of the class y. However, for most \({ \mathop { \text {BN} } }\) classifiers the class has no parents and is made a parent of all attributes. This has the effect that every attribute is in the Markov blanket of the class.
We will assume that the parents of the class attribute constitute an empty set and, therefore, replace parameters characterizing the class attribute from \(\theta _{y^{(j)} | \varPi _0(\mathbf{x}^{(j)})}\) with \(\theta _{y^{(j)}}\). We will also drop the superscript j in equations for clarity.
2.3 A simple framework
It is no exaggeration to say that Eq. 1 has a pivotal role in \({ \mathop { \text {BN} } }\) classification. Let us modify Eq. 1 by introducing an extra set of parameter, say w for every parameter \(\theta \). Let \({\varvec{\theta }}\) and \(\mathbf{w}\), represent the vectors of all \(\theta \) and w parameters. In the following, let us also make a distinction between ‘Fixed’ and ‘Optimized’ parameters—during the optimization process. Parameters that are optimized are referred to as Optimized parameters and parameters that do not change their value during the optimization process are referred to as Fixed. Now, we can write:
Optimizing to compute class-probabilities using Eq. 6, there are several possibilities, of which we will discuss only four in the following:
-
1.
Generative: Initialize \(\mathbf{w}\) with 1 and treat it as fixed parameter. Treat, \({\varvec{\theta }}\) as optimized parameter and optimize it with the generative objective function as given in Eq. 3.
-
2.
Discriminative: Initialize \(\mathbf{w}\) with 1 and treat it as fixed parameter. Treat, \({\varvec{\theta }}\) as an optimized parameter and optimize it with the discriminative objective function as given in Eq. 5. As discussed, this results in adding a normalization term to convert \(\mathrm{P}(y,\mathbf{x})\) in Eq. 6 to \(\mathrm{P}(y|\mathbf{x})\). We denote this ‘discriminative CCBN’ and describe it in detail in Sect. 3.
-
3.
Discriminative: Initialize \(\mathbf{w}\) with 1 and treat it as a fixed parameter. Treat, \({\varvec{\theta }}\) as an optimized parameter and optimize it with the discriminative objective function as given in Eq. 5, but constrain parameter \({\varvec{\theta }}\) to be actual probabilities. We denote this ‘extended CCBN’ and provide a detailed description in Sect. 4.
-
4.
Discriminative: Two step learning. In the first step, initialize \(\mathbf{w}\) with 1 and treat it as a fixed parameter. Treat, \({\varvec{\theta }}\) as an optimized parameter and optimize it with the generative objective function as given in Eq. 3. In the second step, treat \({\varvec{\theta }}\) as a fixed parameter and optimize for \(\mathbf{w}\) using a discriminative objective function. This approach is inspired from the fact that weights \(\mathbf{w}\) in Eq. 6 are set through generative learning, unlike discriminative and extended CCBN, where it is set to one. We denote this ‘weighted CCBN’ and describe it in detail in Sect. 5.
A brief summary of these parameterizations is also given in Table 2.
3 Parameterization 1: Discriminative CCBN model
Logistic regression (LR) is the CCBN model associated to the NB structure optimizing Eq. 2. Typically, LR learns a weight for each attribute-value (per-class). However, one can extend LR by considering all or some subset of possible quadratic, cubic, or higher-order features (Langford et al. 2007; Zaidi et al. 2015). Inspired from Roos et al. (2005), we define discriminative CCBN as:
Definition 2
A discriminative class-conditional Bayesian network model \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) is a CCBN such that Eq. 2 is re-parameterized in form of parameter \({\varvec{\beta }}\) such that \({\varvec{\beta }}= \log {\varvec{\theta }}\) and parameter \({\varvec{\beta }}\) is obtained by maximizing the CLL.
Let us re-define \(\mathrm{P}_{\mathcal {B}}(y|\mathbf{x})\) in Eq. 5 and write it on a per datum basis as:
In light of Definition 2, let us define a parameter \(\beta _{\bullet }\) that is associated with each parameter \(\theta _{\bullet }\) in Eq. 7, such that:
Now Eq. 7 can be written as:
One can see that this has led to the logistic function of the form \(\frac{1}{1+\exp (-\mathbf {\beta }^T\mathbf{x})}\) for binary classification and softmax \(\frac{\exp (-\mathbf {\beta _y}^T\mathbf{x})}{\sum _y' (\exp (-\mathbf {\beta _{y'}}^T\mathbf{x}))}\) for multi-class classification. Such a formulation is a Logistic Regression classifier. Therefore, we can state that a discriminative CCBN model with naive Bayes structure is a (vanilla) logistic regression classifier.
In light of Definition 2, CLL optimized by \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\), on a per-datum-basis, can be specified as:
Now, we will have to rely on an iterative optimization procedure based on gradient-descent. Therefore, let us first calculate the gradient of parameters in the model. The gradient of the parameters in Eq. 9 can be computed as:
for the class parameters. For the other parameters, we can compute the gradient as:
where \(\mathbf {1}\) is the indicator function. Note, that we have used the notation \(\beta _{y:k,x_i:j,\varPi _i:l}\) to denote that class y has the value k, attribute \(x_i\) has the value j and its parents (\(\varPi _i\)) have the value l. If the attribute has multiple parent attributes, then l represents a combination of parent attribute values.
4 Parameterization 2: Extended CCBN model
The name Extended CCBN Model is inspired from Greiner et al. (2005), where the method named Extended Logistic Regression (ELR) is proposed. ELR is aimed at extending LR and leads to discriminative training of \({ \mathop { \text {BN} } }\) parameters. We define:
Definition 3
(Greiner et al. 2005) An extended class-conditional Bayesian network model \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) is a CCBN such that the parameters (\({\varvec{\theta }}\)) satisfy the constraints in Eq. 4 and is obtained by maximizing the CLL in Eq. 5.
Let us re-define \(\mathrm{P}_{\mathcal {B}}(y|\mathbf{x})\) in Eq. 5 on a per-datum-basis as:
Let us consider the case of optimizing parameters associated with the attributes \(\theta _{x_i|y,\varPi _i(\mathbf{x})}\). Parameters associated with the class can be obtained similarly. We will re-write \(\theta _{x_i|y,\varPi _i(\mathbf{x})}\) as \(\theta _{x_i:j|y:k,\varPi _i:l}\) which represents attribute i (\(x_i\)) taking value j, class (y) taking value k and its parents (\(\varPi _i\)) takes value l. Now we can write the gradient as:
Enforcing constraints that \(\sum _{j'} \theta _{x_i:j'|y:k,\varPi _i:l} = 1\), we introduce a new parameters \(\beta \) and re-parameterize as:
It will be helpful if we differentiate \(\theta _{x_i:j'|y:k,\varPi _i:l}\) with respect to \(\beta _{x_i:j|y:k,\varPi _i:l}\) (the use of notation j and \(j'\) will become obvious when we apply the chain rule afterwards), we get:
Applying the chain rule:
we get the gradient of \(\log \mathrm{P}_{\mathcal {B}}(y|\mathbf{x})\) with respect to parameter \(\beta _{x_i:j|y:k,\varPi _i:l}\). Now one can use the transformation of Eq. 13 to obtain the desired parameters of extended CCBN. Note that Eq. 14 corresponds to Eq. 11. The only difference is the presence of the normalization term that is subtracted from the gradient in Eq. 14.
5 Parameterization 3: Combined generative/discriminative parameterization: weighted CCBN model
Inspired from Zaidi et al. (2014), we define a weighted CCBN model as follows:
Definition 4
A weighted conditional Bayesian network model \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) is a CCBN such that Eq. 2 has an extra weight parameter associated with every \(\theta \) such that it is re-parameterized as: \({\varvec{\theta }}^\mathbf{w}\), where parameter \({\varvec{\theta }}\) is learned by optimizing the LL and parameter \(\mathbf{w}\) is obtained by maximizing the CLL.
In light of Definition 4, let us re-define Eq. 2 to incorporate weights as:
The corresponding weighted CLL can be written as:
Note, that Eq. 16 is similar to Eq. 12 except for the introduction of weight parameters. The flexibility to learn parameter \({\varvec{\theta }}\) in a prior generative process of learning greatly simplifies subsequent calculations of \(\mathbf{w}\) in a discriminative search. Since \(\mathbf{w}\) is a free-parameter and there is no sum-to-one constraint, its optimization is simpler than for \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\). The gradient of the parameters in Eq. 16 can be computed as:
for the class y, while for the other parameters:
One can see that Eqs. 17 and 18 correspond to Eqs. 10 and 11. The only difference between them is the presence of the \(\log \theta _{\bullet }\) factor in the \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) case.
5.1 On initialization of parameters
Initialization of the parameters, which sets the starting point for the optimization, is critical to the speed of convergence and will be addressed in this section. Obviously, a better starting point (in terms of CLL), will make the optimization easier and conversely, a worse starting point will make optimization harder. In this paper, we will study two different starting points for the parameters:
-
Initialization with Zeros This is the standard initialization where all the optimized parameters are initialized with 0 (Ripley 1996).
-
Initialization with Generative estimates Given that our approach utilizes generative estimates, a fair comparison with other approaches should study starting from the generative estimates for all approaches. This will correspond to initializing the \({\varvec{\theta }}\) parameter with the generative estimates for Parameterizations 1 and 2 (\({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\)), and initializing the \(\mathbf{w}\) parameter to 1 for Parameterization 3 (\({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\)).
Note that in the initialization with “Zeros” case, only our proposed Weighted CCBN parameterization requires a first (extra) pass over the dataset to compute the generative estimates, while for the initialization with “Generative estimates” case all methods require this pass (when we report training time, we always report the full training time).
5.2 Comment on regularization
\({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) parameterization offers an elegant framework for blending discriminatively and generatively learned parameters. With regularization, one can indeed ‘interpolate’ between the two sets of parameters. Traditionally, one regularizes parameters towards 0 to prevent over-fitting. For example, let us modify Eq. 15 to integrate an L2-regularization:
where \(\mathcal {Z}\) is the normalization constant and \(\lambda \) is the parameter controlling regularization. The new term will penalize large (and heterogeneous) parameter values. Larger \(\lambda \) values will cause the classifier to progressively ignore the data and assign more uniform class probabilities. Alternatively one could penalize deviations from the \({ \mathop { \text {BN} } }\) conditional independence assumption by centering the regularization term at 1 rather than zero. In this case, we can write:
Doing so allows the regularization parameter \(\lambda \) to be used to ‘pull’ the dicriminative estimates toward the generative ones. A very small value of \(\lambda \) results in optimized parameter \(\mathbf{w}\) dominating the determination of \(\mathrm{P}(y|\mathbf{x})\), whereas, a very large value of \(\lambda \) pulls \(\mathbf{w}\) towards 1 and, therefore, the fixed parameters will dominate the class-conditional probabilities. Regularization for \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) remains an area for future research, but we conjecture that one can tune a value of \(\lambda \) (for example through cross-validation) to attain better performance than can be achieved by either generative or discriminative parameters alone. Once could also interpret the regularization parameter as controlling the amount of independence violation between the discriminative and generative models.
5.3 Optimizing discriminative/generative parameterization
There are great advantages in optimizing an objective function that is convex. The convexity of the three discriminative parameterizations that we have discussed depends on the underlying structure of the CCBN (\({\mathcal {M}^{{\mathcal {B}}^*}}\)). From Roos et al. (2005), it follows that optimizing a CCBN parameterized by either \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) or \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) leads to a convex optimization problem if and only if the structure has no immoral nodes. In other words, the optimization problem is convex if and only if parents of all the nodes are are connected with each other. This constraint is true for a number of popular \({ \mathop { \text {BN} } }\) classifiers including NB, TAN and KDB (\(K=1\)), but not true for general \({ \mathop { \text {BN} } }\) or for KDB structures with \(K > 1\). Therefore, in this work, we have used only limited \({ \mathop { \text {BN} } }\) structures such as NB, TAN and KDB (\(K=1\)). Investigation of the application of our approach to more complex moral structures is a promising topic for future work. We note in passing, that a similar two step discriminative parameterization has also been shown to be effective for the non-convex objective function mean-square-error (Zaidi et al. 2016).
5.4 Nested parameterizations
One can see that learning \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) models can lead to a large number of parameters that needs to be optimized discriminatively, even on moderate size datasets. One can, however, nest these parameters. The idea is to exploit relationships between parameters so that the number of parameters that need to be optimized are reduced significantly. Figure 1 depicts four levels of parameter nesting. The first level entails learning a parameter for each attribute. The second level entails learning a parameter for every attribute-value. The next level learns a parameter for every attribute-value-per-class-value. The final level (Level 4) is the most comprehensive case. It entails learning a parameter for every attribute-value-per-class-per-parent-value.
Depiction of various levels in parameter nesting, along with number of parameters (m) to be optimized at each level. Note that only one node per level is expanded, for illustration. Attribute \(X_1\) takes values \(\{a,b,c\}\) and takes class \(Y = \{ 0,1\}\) and attribute \(X_2 = \{ d,e,f \}\) as parents
Nesting as shown in Fig. 1, though effective, is not very intuitive for \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\). For example, doing a logistic regression by learning a parameter associated only with the attributes will result in optimizing fewer parameters but might not be effective in terms of classification accuracy. However, the hierarchy of models applies naturally to \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\). \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) incorporates learning initial parameters by optimizing the LL objective function. Therefore, the searched parameters optimized in the second step can be nested effectively. For example, Level 1 weighting in Fig. 1 can be seen as alleviating the conditional attribute independence assumption (CAIA) between attributes. Similarly, Level 2 will have the effect of binarizing each attribute, and alleviating CAIA between new attributes. In the following we will derive the respective gradients for each level from the most comprehensive case of Level 4.
Gradients for Level 4 are given in Eqs. 17 and 18. Level 3 corresponds to learning a weight-per-attribute-value-per-class. The weight vector in this case will be of the size \(m = \sum _i (|Y| \times | X_{i} |)\). The gradients with respect to new weight vectors can be obtained in the following way:
Level 2 weighting corresponds to learning a weight-per-attribute-value. We can compute the gradient with respect to the weight vector of size \(m = \sum _i | X_i |\), as:
Similarly, learning a weight-per-attribute leads to a weight vector of size m and can its gradients can be obtained as:
Now, one can control the bias and variance of the classifier by selecting between three different levels of parameterization with ever greater model complexity.
5.5 Discussion
5.5.1 Pre-conditioning: why is our technique helpful?
It can be seen that \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) results in re-scaling of \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) parameterization. What is the effect of this re-scaling on the model? Since there is no closed-form solution, we optimize the CLL with first-order gradient-descent methods, such as gradient descent, conjugate gradient, quasi-Newton (L-BGFS) or Stochastic Gradient Descent. These are all affected by scaling.Footnote 1 We use the generative estimates as an effective pre-conditioning method.
A pre-conditioner converts an ill-conditioned problem into a better conditioned one, such that the gradient of the objective function is uniform across all dimensions. A better conditioned optimization problem has a better convergence profile. This is because if different parameters have significantly different “influence” on the objective function, then the gradient does not point directly towards the minimum that is the objective of the optimization process. We illustrate this in Fig. 2 where we show the contour plot of the CLL for different \(\beta \). We can see that when the CLL has an ‘elliptical’ shape with respect to the parameters, then the gradient is not oriented directly towards the objective and each step makes only partial progress in the true direction of the final objective. Our re-scaling improves the orientation of the gradient speeding convergence.
Note that it is the relative scaling of the axes that affects the orientation of the gradient. Isotropic scaling (that is, scaling all axes uniformly) has no effect on convergence.
To further demonstrate our point, we perform a simple experiment with synthetic data that we generate so that the CLL is more or less “elliptical”. We use with three binary features and two class values. We sample the covariates randomly and uniformly and use a simple logistic regression model, which corresponds to our framework using Naive Bayes as the BN structure. The class distribution is given by
By increasing the value of \(\alpha \), we increase the elongation of the CLL space. When \(\alpha = 0\), the three features contribute uniformly to the class prediction and it is a well-conditioned problem. We can then expect pre-conditioning to have little to no influence on the convergence. As \(\alpha \rightarrow 1\), the problem becomes very ill-conditioned. On such problems, pre-conditioning will have greatest effect.
We compare the convergence profile of vanilla LR (discriminative CCBN) and our pre-conditioned weighted CCBN with Naive Bayes structure (which is associated to a LR model) by varying \(\alpha \) from 0 to 1. For each dataset 10000 data points were generated. We report the convergence results in Fig. 3. These confirm the explanation given above. The benefit of our technique progressively increases as the relative influence of the covariates on the class increases. We will show in Sect. 7 that this is the case for the vast majority of real-world datasets.
5.5.2 Is this an over-parametrised model?
It can be seen that the \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) parameterization is based on Eq. 6—which is over-parameterized in the sense that there are twice as many parameters specifying the likelihood as would strictly be necessary. The question is: do we benefit from having both \(\mathbf{w}\) and \({\varvec{\theta }}\) parameters? In this section, we will discuss the implications of introducing \(\mathbf{w}\) parameters to the following vanilla (per-datum) likelihood (on which \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) is based on):
If one goal of weighted CCBN is to combine generative and discriminative learning by using over-parameterized likelihood in Eq. 6, one could do the following two-step learning. In step 1, one can learn the \({\varvec{\theta }}\) by optimizing a generative objective function, and in the second step, optimize a discriminative objective function but initialize the \({\varvec{\theta }}\) parameters with the parameters that were obtained in step 1. In fact, this should be a recommended procedure to speed-up discriminative training (for discriminative CCBN and extended CCBN) as it is often effective in practice. However, one should notice that in this case, the discriminative learning model does start from the estimates of parameters that were obtained from generative learning, but once an iterative step is taken for discriminative learning, the generative estimates are lost, and have no further influence on the discriminative learning process.
6 Related work
There have been several comparative studies of discriminative and generative structure and parameter learning of Bayesian networks (Greiner and Zhou 2002; Grossman and Domingos 2004; Pernkopf and Bilmes 2005). In all these works, generative parameter training is the estimation of parameters based on empirical estimates whereas discriminative training of parameters is actually the estimation of the parameters of CCBN models such as \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) or \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\). The \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) model was first proposed in Greiner and Zhou (2002). Our work differs from these previous works as our goal is to highlight different parameterization of CCBN models and investigate their inter-relationship. Particularly, we are interested in the learning of parameters corresponding to a weighted CCBN model that leads to faster discriminative learning.
An approach for discriminative learning of the parameters of \({ \mathop { \text {BN} } }\) based on discriminative computation of pseudo-frequencies from the data is presented in Su et al. (2008). Discriminative Frequency Estimates (DFE) are computed by injecting a discriminative element to generative computation of the probabilities. During the pseudo-frequencies computation process, rather than using empirical frequencies, DFE estimates how well the current classifier does on each data point and then updates the frequency tables only in proportion to the classifier’s performance. For example, they propose a simple error measure, as: \(L(\mathbf{x}) = \mathrm{P}(y|\mathbf{x}) - \hat{\mathrm{P}}(y|\mathbf{x})\), where \(\mathrm{P}(y|\mathbf{x})\) is the true probability of class y given the datum \(\mathbf{x}\), and \(\hat{\mathrm{P}}(y|\mathbf{x})\) is the predicted probability. The counts are updated as: \(\theta _{ijk}^{t+1} = \theta _{ijk}^{t} + L(\mathbf{x})\). Several iterations over the dataset are required. The algorithm is inspired from Perceptron based training and is shown to be an effective discriminative parameter learning approach.
7 Empirical results
In this section, we compare and analyze the performance of our proposed algorithms and related methods on 72 natural domains from the UCI repository of machine learning (Frank and Asuncion 2010). The experiments are conducted on the datasets described in Table 3.
There are a total of 72 datasets, 41 datasets with less than 1000 instances, 21 datasets with between 1000 and 10000 instances, and 11 datasets with more than 10000 instances. Each algorithm is tested on each dataset using 5 rounds of 2-fold cross validation. 2-fold cross validation is used in order to maximize the variation in the training data from trial to trial, which is advantageous when estimating bias and variance. Note that the source code with running instructions is provided as a supplementary material to this paper.
We compare four metrics: 0–1 Loss, RMSE, Bias and Variance. The reason for performing bias/variance estimation is to investigate if optimizing a discriminative function leads to a lower bias classifier or not. There are a number of different bias-variance decomposition definitions. In this research, we use the bias and variance definitions of Kohavi and Wolpert (1996) together with the repeated cross-validation bias-variance estimation method proposed by Webb (2000). Kohavi and Wolpert (1996) define bias and variance as follows:
and
The reason for reporting 0–1 Loss and RMSE is to investigate if the proposed parameterization \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) leads to a comparable performance to \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) parameterizations and also to determine how much performance gain is achieved over generative learning. We will also evaluate parameterizations in terms of training time (measured in seconds) and number of iterations it takes each parameterization to converge.
We report Win–Draw–Loss (W–D–L) results when comparing the 0–1 Loss, RMSE, bias and variance of two models. A two-tail binomial sign test is used to determine the significance of the results. Results are considered significant if \(p \le 0.05\). Significant results are shown in bold font in the table.
We report results on two categories of datasets. The first category, labeled All, consists of all datasets in Table 3. The second category, labeled Big, consists of datasets that have more than 10000 instances. The reason for splitting datasets into two categories is to show explicitly the effectiveness of our proposed optimization on bigger datasetsFootnote 2. It is only on big datasets, that each iteration is expensive and, therefore, any technique that leads to faster and better convergence is highly desirable. Note, that we do not expect the three discriminative parameterizations \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) to differ in their prediction accuracy. That is, we should expect a similar spread of both 0–1 Loss and RMSE values. However, we should be interested in each parameterization’s convergence profile and the training time.
Numeric attributes are discretized using the Minimum Description Length (MDL) discretization method (Fayyad and Irani 1992). A missing value is treated as a separate attribute value and taken into account exactly like other values.
Optimization is done with L-BFGS (Byrd et al. 1995) using the original implementation available at http://users.eecs.northwestern.edu/~nocedal/lbfgsb.html. Following standard procedures (Zhu et al. 1997), the algorithm terminates when improvement in the objective function, given by \(\frac{(f_t - f_{t+1})}{\max \{ |f_t|, |f_{t+1}|,1\}}\), drops below \(10^{-32}\), or the number of iterations exceeds \(10^4\).
Comparative scatter of results for \({ \mathop { \text {NB}^{\mathrm{w}}}}\), \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\). \({ \mathop { \text {NB}^{\mathrm{w}}}}\) is on the X-axis whereas \({ \mathop { \text {NB}^{\mathrm{d}}}}\) (red-cross) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\) (green-triangle) are on the Y-axis. For any points above the diagonal line \({ \mathop { \text {NB}^{\mathrm{w}}}}\) wins
We experiment with three Bayesian network structures that is: naive Bayes (NB), Tree-Augmented naive Bayes (TAN) (Friedman et al. 1997) and k-Dependence Bayesian network (KDB) with \(K=1\) (Sahami 1996). Naive Bayes, is a well-known classifier which is based on the assumption that when conditioned on the class, attributes are independent. Tree-Augmented Naive Bayes augments the NB structure by allowing each attribute to depend on at most one non-class attribute. It relies on an extension of the Chow-Liu tree (Chow and Liu 1968), that utilizes conditional mutual information (between pairs of attributes given the class) to find a maximum spanning tree over the attributes in order to determine the parent of each. Similarly, in KDB, each attribute takes k attributes plus the class as its parents. The attributes are selected based on their mutual information with the class. Then, the parent of an attribute i is chosen that maximizes the conditional mutual information of attribute i and parent j given the class that is: \(\text {argmax}_j \text {CMI}(X_i,X_j | Y)\).
We denote \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) with naive Bayes structure as \({ \mathop { \text {NB}^{\mathrm{w}}}}\), \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\) respectively. With TAN structure, \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) are denoted as \({ \mathop { \text {TAN}^{\mathrm{w}}}}\), \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\). With KDB (\(K=1\)), \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) are denoted as \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\), \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\).
As discussed in Sect. 5.1, we initialize the parameters to the log of the MAP estimates (or parameters optimized by generative learning). The following naming convention is used in the results:
-
The ‘(I)’ in the label represents this initialization
-
An absence of ‘(I)’ means the parameters are initialized to zero.
7.1 NB structure
Comparative scatter plots on all 72 datasets for 0–1 Loss, RMSE and training time values for \({ \mathop { \text {NB}^{\mathrm{w}}}}\), \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\) are shown in Fig. 4. Training time plots are on the log scale. The plots are shown separately for Big datasets. It can be seen that the three parameterizations have a similar spread of 0–1 Loss and RMSE values, however, \({ \mathop { \text {NB}^{\mathrm{w}}}}\) is greatly advantaged in terms of its training time. We will see in Sect. 7.4 that this computational advantage arises due to the desirable convergence property of \({ \mathop { \text {NB}^{\mathrm{w}}}}\). Given that \({ \mathop { \text {NB}^{\mathrm{w}}}}\) achieves equivalent accuracy with much less computation indicates that it is a more effective parameterization than \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\). Slight variation in the accuracy of three discriminative parameterizations (that is 0–1 Loss and RMSE performance) on All datasets is due to the numerical instability of the solver use for optimization. The difference mainly arises on very small datasets. It can be seen that on big datasets, the three parameterizations result in the same accuracy.
The geometric means of the 0–1 Loss and RMSE results are shown in Fig. 5. It can be seen that the three discriminative parameterizations, especially on Big datasets, has much better performance (both 0–1 Loss and RMSE) than the generative learning.
The training time comparison is given in Fig. 6a. Note that the training time is measured in seconds and is plotted on the log scale. It can be seen that in terms of the training time, the three discriminative parameterizations are many orders of magnitude slower than plain naive Bayes. To show the comparison of the training time of \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) only, we normalize the results with respect to \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\). Results are shown in Fig. 6b. It can be seen that \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) is almost twice as slow as compared to \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\), whereas, \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) is order of magnitude slower that \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\).
a Geometric mean of training time for NB, \({ \mathop { \text {NB}^{\mathrm{w}}}}\), \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\) on All and Big datasets. Results are normalized with respect to NB. b Geometric mean of training time for \({ \mathop { \text {NB}^{\mathrm{w}}}}\), \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\) on All and Big datasets. Results are normalized with respect to \({ \mathop { \text {NB}^{\mathrm{w}}}}\)
Comparative scatter of results for \({ \mathop { \text {TAN}^{\mathrm{w}}}}\), \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\). \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) is on the X-axis whereas \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) (red-cross) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) (green-triangle) are on the Y-axis. For any points above the diagonal line \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) wins
7.2 TAN structure
Figure 7 shows the comparative spread of 0–1 Loss, RMSE and training time in seconds of \({ \mathop { \text {TAN}^{\mathrm{w}}}}\), \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) on All and Big datasets. A trend similar to that of NB can be seen. With a similar spread of 0–1 Loss and RMSE among the three parameterizations, training time is greatly improved for \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) when compared with \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\). Note, as pointed out before, that minor variation in the performance of three discriminative parameterizations is due to the numerical issues with-in the solver on some small datasets. On big datasets, one can see a similar spread of 0–1 Loss and RMSE.
The geometric means of the 0–1 Loss and RMSE results are shown in Fig. 8. It can be seen that \({ \mathop { \text {TAN}^{\mathrm{d}}}}\), \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{w}}}}\), on average results in much better accuracy than generative model (TAN).
A comparison of the training time is shown in Fig. 9a. It can be seen that, like NB, training time of the discriminative methods is orders of magnitude longer than that of generative learning. Note, the training time of discriminative learning also includes the structure learning process. We also show the comparison of the training time of \({ \mathop { \text {TAN}^{\mathrm{d}}}}\), \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) in Fig. 9b. Like, NB, it can be seen that \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) is almost twice as fast as \({ \mathop { \text {TAN}^{\mathrm{d}}}}\), whereas, \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) is orders of magnitude slower than \({ \mathop { \text {TAN}^{\mathrm{w}}}}\).
a Geometric mean of training time for TAN, \({ \mathop { \text {TAN}^{\mathrm{w}}}}\), \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) on All and Big datasets. Results are normalized with respect to NB. b Geometric mean of training time for \({ \mathop { \text {TAN}^{\mathrm{w}}}}\), \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) on All and Big datasets. Results are normalized with respect to \({ \mathop { \text {TAN}^{\mathrm{w}}}}\)
Comparative scatter of results for \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\), \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\). \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) is on the X-axis whereas \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) (red-cross) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\) (green-triangle) are on the Y-axis. For any points above the diagonal line \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) wins
7.3 KDB (\(K=1\)) structure
Figure 10 shows the comparative spread of 0–1 Loss, RMSE and training time in seconds of \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\), \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\) on All and Big datasets. Like NB and TAN, it can be seen that a similar spread of 0–1 Loss and RMSE is present among the three parameterizations of discriminative learning. Similarly, it can be seen that training time is greatly improved for \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) when compared with \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\).
Geometric average of the 0–1 Loss and RMSE results are shown in Fig. 11. It can be seen that the three discriminative parameterizations have better 0–1 Loss and RMSE than generative learning (KDB-1).
A comparison of the training time is given in Fig. 12a. Note, the training time of discriminative methods also includes the time of structure learning. It can be seen that discriminative learning leads to a significantly longer training time than generative learning. We compare the training time of \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\), \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) in Fig. 12b. Like, NB and TAN structure, it can be seen that \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) is faster than both \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\).
a Geometric mean of training time for KDB1, \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\), \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\) on All and Big datasets. Results are normalized with respect to NB. b Geometric mean of training time for \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\), \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\) on All and Big datasets. Results are normalized with respect to \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\)
7.4 Convergence analysis
A comparison of the convergence of Negative Log-Likelihood (NLL) of the three parameterizations on some sample datasets with NB, TAN and KDB (\(K=1\)) structure is shown in Figs. 13 and 14.
As discussed in Sect. 5.1, in Fig. 13, parameters are initialized to zero, whereas, in Fig. 14, parameters are initialized to the log of the MAP estimates. It can be seen that for all three structures and for both initializations, \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) not only converges faster but also reaches its asymptotic value much quicker than the \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\). The same trend was observed on all 72 datasets. A comparison on many more datasets is given in Figs. 18 and 19 in Appendix 2.
Comparison of rate of convergence on the four biggest datasets for NB, TAN and KDB (\(K=1\)) (right column) structures. The X-axis is on log scale. Parameters are initialized to zero. Note, the first iteration is actually NLL before the start of optimization. It can be seen that the three parameterizations start from the same point in the space
Comparison of rate of convergence on the four biggest datasets for NB, TAN and KDB (\(K=1\)) (right column) structures. The X-axis is on log scale. Parameters are initialized to the log of the MAP estimates. Note, the first iteration is actually NLL before the start of optimization. It can be seen that the three parameterizations start from the same point in the space
Number of iterations (after 5 iterations), it takes \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\) to reach NLL that \({ \mathop { \text {NB}^{\mathrm{w}}}}\) achieved after 5 iterations. If NLL of \({ \mathop { \text {NB}^{\mathrm{w}}}}\) is less that that of \({ \mathop { \text {NB}^{\mathrm{d}}}}\) or \({ \mathop { \text {NB}^{\mathrm{e}}}}\), (negative of the) number of iterations it takes \({ \mathop { \text {NB}^{\mathrm{w}}}}\) to reach that of \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and that of \({ \mathop { \text {NB}^{\mathrm{e}}}}\) are plotted with negative sign
Number of iterations (after 5 iterations), it takes \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) to reach NLL that \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) achieved after 5 iterations. If NLL of \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) is less that that of \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) or \({ \mathop { \text {TAN}^{\mathrm{e}}}}\), (negative of the) number of iterations it takes \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) to reach that of \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and that of \({ \mathop { \text {TAN}^{\mathrm{e}}}}\) are plotted with negative sign
Number of iterations (after 5 iterations), it takes \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\) to reach NLL that \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) achieved after 5 iterations. If NLL of \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) is less that that of \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) or \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\), (negative of the) number of iterations it takes \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) to reach that of \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and that of \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\) are plotted with negative sign
To quantify how much \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) is faster than the other two parameterizations, we plot a histogram of the number of iterations it takes \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) after five iterations to reach the negative log-likelihood that \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) achieved in the fifth iteration. If the three parameterizations follow similar convergence, one should expect many zeros in the histogram. Note that if after the fifth iteration, NLL of \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) is greater than that of \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\), we we plot the negative of the number of iterations it takes \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) to reach the NLL of \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\). Similarly, if after the fifth iteration, NLL of \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) is greater than that of \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\), we we plot the negative of the number of iterations it takes \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) to reach the NLL of \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\). Figures 15, 16 and 17 show these histogram plots for NB, TAN and KDB (\(K=1\)) structure respectively. It can be seen that \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) (with all three structures) achieves a NLL that otherwise, will take on average 10 more iterations over the data for \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and 15 more iterations for \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\). This is an extremely useful property of \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) especially for big data where iterating through the dataset is expensive.
7.5 Comparison with MAP
The purpose of this section is to compare the performance of the discriminative learning with that of generative learning. In Table 4, we compare the performance of \({ \mathop { \text {NB}^{\mathrm{w}}}}\) with NB (i.e., naive Bayes with MAP estimates of probabilities), \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) with TAN (i.e., TAN with MAP estimates of probabilities) and \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) with KDB (\(K=1\)) (i.e., KDB with MAP estimates of probabilities). We use \({ \mathop { \text {NB}^{\mathrm{w}}}}\), \({ \mathop { \text {TAN}^{\mathrm{w}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\) as a representative of discriminative learning - since \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) have similar 0–1 Loss and RMSE profile. It can be see that the discriminative learning of parameters has significantly lower bias but higher variance. On big datasets, it can be seen that discriminative learning results in much better 0–1 Loss and RMSE performance.
Note that though discriminative learning (optimizes the parameters characterizing CCBN) has better 0–1 Loss and RMSE performance than generative learning (optimizing joint probability),—generative learning has the advantage of being extremely fast as it incorporates counting of sufficient statistics from the data. Another advantage of generative learning is its capability of back-off in case a certain combination does not exist in the data. For example, if TAN or KDB classifiers have not encountered a \({<}{} \texttt {feature-value, parent-value, class-value}{>}\) combination at training time they can resort back to \({<}{} \texttt {feature-value, class-value}{>}\) at testing time. For instance TAN classifier can step back to NB and NB can step back to class prior probabilities. Such elegantly back-tracking is missing from discriminative learning. If a certain combination does not exist in the data, parameters associated to that combination will not be optimized and will remain fixed to the initialized value (for example 0). A discriminative classifier will have no way of handling unseen combinations but to ignore them if they occur in the testing data. How to incorporate such hierarchical learning with discriminative learning is the goal of future research as will be discussed in Sect. 8.
8 Conclusion and future work
In this paper, we propose an effective parameterization of \({ \mathop { \text {BN} } }\). We present a unified framework for learning the parameters of Bayesian network classifiers. We formulate three different parameterizations and compare their performance in terms of 0–1 Loss, RMSE and training time each parameterization took to converge. We show with NB, TAN and KDB structures that the proposed weighted discriminative parameterization has similar 0–1 Loss and RMSE to the other two but significantly faster convergence. We also show that it not only has faster convergence but it also asymptotes to its global minimum much quicker than the other two parameterizations. This is desirable when learning from huge quantities of data with Stochastic Gradient Descent (SGD). It is also shown that discriminative training of \({ \mathop { \text {BN} } }\) classifiers also leads to lower bias than the generative parameter learning.
We plan to conduct following future work as the result of this study:
-
The three parameterizations presented in this work learn a weight for each attribute-value-per-class-value-per-parent-values. As discussed in Sect. 5.4, contrary to \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) parameterization can generalize parameters. For example, once MAP estimates of probabilities are learned, one can learn a weight: (a) for each attribute only (i.e., same weight for all attribute-values, for all class values and for all parent values), (b) for each attribute-value only, (c) for each attribute-value-per-class-value, (d) for each attribute-value-per-class-value-per-parent, etc. Such parameter generalization could offer additional speed-up of the training and is a promising avenue for future research.
-
Handling combinations of \(\langle \texttt {feature-value}, \texttt {parent-value}, \texttt {class-value}\rangle \) that have not been seen at training time is one of the weaker properties of discriminative learning. We plan to design an hierarchical algorithm of discriminative learning that can learn lower-level discriminative weights and can back-off from higher levels if a combination is not observed in the training data.
-
We plan to conduct an extended analysis of \({ \mathop { \text {BN} } }\) models that can capture higher-order interactions. Because the CLL is not convex for most of these models (Roos et al. 2005), it falls outside the scope of this paper. This does, however, suggest inviting avenues for big data research, in which context low-bias classifiers are required.
9 Code and datasets
All the datasets used in this paper are in the public domain and can be downloaded from Frank and Asuncion (2010). Code with running instructions can be download from https://github.com/nayyarzaidi/EBNC.git.
Notes
Note that second-order algorithms such as the Newton method are not affected by scaling, but they are often computationally impractical because they require computation and inversion of the Hessian at each step.
By big, we mean datasets that have large number of instances, rather than large number of features
References
Buntine, W. (1994). Operations forlearning with graphical models. Journal of Artificial Intelligence Research, 2, 159–225.
Byrd, R., Lu, P., & Nocedal, J. (1995). A limited memory algorithm for bound constrained optimization. SIAM Journal on Scientific and Statistical Computing, 16(5), 1190–1208.
Carvalho, A., Roos, T., Oliveira, A., & Myllymaki, P. (2011). Discriminative learning of Bayesian networks via factorized conditional log-likelihood. Journal of Machine Learning Research.
Chow, C., & Liu, C. (1968). Approximating discrete probability distributions with dependence trees. IEEE Transactions on Information Theory, 14(3), 462–467.
Demšar, J. (2006). Statistical comparisons of classifiers over multiple data sets. Journal of Machine Learning Research, 7, 1–30.
Fayyad, U. M., & Irani, K. B. (1992). On the handling of continuous-valued attributes in decision tree generation. Machine Learning, 8(1), 87–102.
Frank, A., & Asuncion, A. (2010). UCI machine learning repository. http://archive.ics.uci.edu/ml.
Friedman, N., Geiger, D., & Goldszmidt, M. (1997). Bayesian network classifiers. Machine Learning, 29(2), 131–163.
Greiner, R., & Zhou, W. (2002). Structural extension to logistic regression: Discriminative parameter learning of belief net classifiers. In Annual national conference on artificial intelligence (AAAI), pp. 167–173.
Greiner, R., Su, X., Shen, B., & Zhou, W. (2005). Structural extensions to logistic regression: Discriminative parameter learning of belief net classifiers. Machine Learning, 59, 297–322.
Grossman, D., & Domingos, P. (2004). Learning Bayesian network classifiers by maximizing conditional likelihood. In ICML.
Heckerman, D., & Meek, C. (1997). Models and selection criteria for regression and classification. In International conference on uncertainty in artificial intelligence.
Jebara, T. (2003). Machine Learning: Discriminative and Generative. Berlin: Springer.
Kohavi, R., & Wolpert, D. (1996). Bias plus variance decomposition for zero-one loss functions. In ICML (pp. 275–283).
Langford, J., Li, L., & Strehl, A. (2007). Vowpal wabbit online learning project. https://github.com/JohnLangford/vowpal_wabbit/wiki.
Martinez, A., Chen, S., Webb, G. I., & Zaidi, N. A. (2016). Scalable learning of Bayesian network classifiers. Journal of Machine Learning Research, 17, 1–35.
Ng, A., & Jordan, M. (2002). On discriminative vs. generative classifiers: A comparison of logistic regression and naive bayes. In Advances in neural information processing systems.
Pernkopf, F., & Bilmes, J. (2005). Discriminative versus generative parameter and structure learning of Bayesian network classifiers. In ICML.
Pernkopf, F., & Bilms, J. A. (2010). Efficient heuristics for discriminative structure learning of Bayesian network classifiers. Journal of Machine Learning Research, 11, 2323–2360.
Pernkopf, F., & Wohlmayr, M. (2009). On discriminative parameter learning of Bayesian network classifiers. In ECML PKDD.
Ripley, B. D. (1996). Pattern Recognition and Neural Networks. Cambridge: Cambridge University Press.
Roos, T., Wettig, H., Grünwald, P., Myllymäki, P., & Tirri, H. (2005). On discriminative Bayesian network classifiers and logistic regression. Machine Learning, 59(3), 267–296.
Rubinstein, Y. D., & Hastie, T. (1997). Discriminative vs informative learning. In AAAI.
Sahami, M. (1996). Learning limited dependence bayesian classifiers. In Proceedings of the second international conference on knowledge discovery and data mining (pp. 335–338).
Su, J., Zhang, H., Ling, C., & Matwin, S. (2008). Discriminative parameter learning for Bayesian networks. In ICML.
Webb, G. I. (2000). Multiboosting: A technique for combining boosting and wagging. Machine Learning, 40(2), 159–196.
Webb, G. I., Boughton, J., Zheng, F., Ting, K. M., & Salem, H. (2012). Learning by extrapolation from marginal to full-multivariate probability distributions: decreasingly naive Bayesian classification. Machine Learning, 86(2), 233–272.
Zaidi, N. A., Carman, M. J., Cerquides, J., & Webb, G. I. (2014). Naive-bayes inspired effective pre-conditioners for speeding-up logistic regression. In IEEE international conference on data mining.
Zaidi, N. A., Cerquides, J., Carman, M. J., & Webb, G. I. (2013). Alleviating naive Bayes attribute independence assumption by attribute weighting. Journal of Machine Learning Research, 14, 1947–1988.
Zaidi, N. A., Petitjean, F., & Webb, G. I. (2016). Preconditioning an artificial neural network using naive bayes. In Proceedings of the 20th Pacific–Asia conference on knowledge discovery and data mining (PAKDD).
Zaidi, N. A., Webb, G. I., Carman, M. J., & Petitjean, F. (2015). Deep Broad Learning—Big models for big data. arXiv:1509.01346.
Zhu, C., Byrd, R. H., & Nocedal, J. (1997). LBFGSB, Fortran routines for large scale bound constrained optimization. ACM Transactions on Mathematical Software, 23(4), 550–560.
Acknowledgements
This research has been supported by the Australian Research Council (ARC) under Grant DP140100087. This material is based upon work supported by the Air Force Office of Scientific Research, Asian Office of Aerospace Research and Development (AOARD) under Award Numbers FA2386-15-1-4007 and FA2386-16-1-4023. The authors would like to thank Reza Haffari and Ana Martinez for helpful discussion during the course of this research. Authors would like to acknowledge Jeusus Cerquides for his extremely useful ideas and suggestions that shaped this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editors: Thomas Gärtner, Mirco Nanni, Andrea Passerini, and Celine Robardet.
Appendices
Appendix 1: Proof of Theorem 1
Let us use Lagrange multipliers for constraints in Eq. 4 to be placed in Eq. 3. Now, we can maximize the resulting objective function:
by first computing its derivative as:
and then setting it to zero. This will lead to
where \(N_{x_i,y,\varPi _i(\mathbf{x})}\) is the empirical count of instances with attribute i taking value \(x_i\), class taking value y and parents taking value \(\varPi _i (\mathbf{x})\). Placing \(\theta _{x_i|\varPi _i(\mathbf{x})}\) value in Eq. 4, we get:
which implies: \(\lambda _i = \sum _{x_i \in \mathcal {X}_i} \sum _{j=1}^{N} N_{x_i,y,\varPi _i(\mathbf{x})}\). Therefore, \(\lambda _i = N_{y,\varPi _i(\mathbf{x})}\). Hence we can write:
This equals empirical estimates of probabilities from the data: \(\mathrm{P}_\mathcal {D}(x_i | \varPi _i(\mathbf{x}))\).
Appendix 2: Convegence curves
Continued from Sect. 7.4, in this section, we present some more results to compare the convergences of three discriminative parameterizations. In Fig. 18, we initialize the parameterizations with the generative estimates, whereas, in Fig. 19, parameters are initialized to zero.
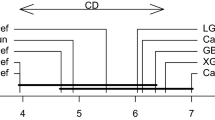
Significance testing of Training time (a) and No. of Iterations (b) to converge for three parameterizations \({ \mathop { \text {NB}^{\mathrm{w}}}}\), \({ \mathop { \text {NB}^{\mathrm{d}}}}\) and \({ \mathop { \text {NB}^{\mathrm{e}}}}\), with Friedman and Nemenyi test on All datasets. Ranks are different according to Friedman test and, therefore, null-hypothesis is rejected. (Nemenyi) Post-hoc test is powerful
Significance testing of Training time (a) and No. of Iterations (b) to converge for three parameterizations \({ \mathop { \text {TAN}^{\mathrm{w}}}}\), \({ \mathop { \text {TAN}^{\mathrm{d}}}}\) and \({ \mathop { \text {TAN}^{\mathrm{e}}}}\), with Friedman and Nemenyi test on All datasets. Ranks are different according to Friedman test and, therefore, null-hypothesis is rejected. (Nemenyi) Post-hoc test is powerful
Significance testing of Training time (a) and No. of Iterations (b) to converge for three parameterizations \({ \mathop { \text {KDB-1}^{\mathrm{w}}}}\), \({ \mathop { \text {KDB-1}^{\mathrm{d}}}}\) and \({ \mathop { \text {KDB-1}^{\mathrm{e}}}}\), with Friedman and Nemenyi test on All datasets. Ranks are different according to Friedman test and, therefore, null-hypothesis is rejected. (Nemenyi) Post-hoc test is powerful
Appendix 3: Training time significance test
Let us discuss the significance of the training time for three discriminative parameterizations using Friedman and Nemenyi tests. Following procedure is taken to generate the results:
-
We have three algorithms to compare that is: \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\), \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\), therefore, \(k = 3\).
-
We compare the results on 72 datasets, therefore, \(N = 72\).
-
Friedman test rank each algorithm for each dataset separately. In case of ties, it uses average ranks.
-
If \(r_i^j\) is the rank of algorithm j on i-th dataset, average rank for each algorithm compared are computed as: \(R_j = \frac{1}{N} \sum _i r_i^j\).
-
We state:
-
Null hypothesis—Algorithms are equivalent and, therefore, ranks should be equal. Mean rank is 2.5.
-
p value—probability of getting ranks \(R_j\) if null-hypothesis as stated in previous point is true.
-
-
Compute the Friedman statistics:
$$\begin{aligned} \chi ^2_F = \frac{12N}{k(k+1)} \left[ \sum _j R_j^2 - \frac{k(k+1)^2}{4} \right] , \end{aligned}$$to determine if the measure ranks are significantly different from the mean rank of 2.5 (under null hypothesis).
-
If the p value is \({\le }0.05\), we reject the null hypothesis and proceed with the post-hoc test.
-
We use the Nemenyi test which states that the performance of two algorithms is significantly different if the corresponding average ranks differ by at least the critical difference (CD) of:
$$\begin{aligned} {\text {CD}} = q_{\alpha } \sqrt{\frac{k(k+1)}{6N}}, \end{aligned}$$where \(q_\alpha \) in our experiments is 2.3430 as \(k = 3\).
-
If the difference between top rank and the bottom rank is less than the CD, we conclude pos-hoc test to be not powerful.
-
Otherwise [following the graphical representation of Demšar (2006)], we plot the ranks (along with the name of algorithm) on a horizontal line. Algorithms are connected by a line if their differences are not significant. We also show the CD on the same scale to highlight the significance of the difference of two ranks.
We show the significance test using Friedman and Nemenyi test on All datasets in terms of training time and no. of iterations it takes each algorithm to converge in Figs. 20, 21 and 22. It can be seen that for all three structures that is NB, TAN and KDB-1, \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) is rank lower than \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) both in terms of training time and no. of iterations.
For NB and TAN, \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) has significantly better training time and converges in far fewer iterations than the other two. However, for KDB-1 structure, the difference is not significant between \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) and \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\).
Appendix 4: Convergence significance test
We compare the NLL obtained by each parameterization at fifth (denoted as NLL (5)), tenth (denoted as NLL (10)) and fiftieth (denoted as NLL (50)) iteration and presented results in terms of win–draw–loss in Table 5 for \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) versus \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) and for \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) versus \({\mathcal {M}_{\text {e}}^{{\mathcal {B}}^*}}\) in Table 6. It can be seen from the two tables, that \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) wins significantly against \({\mathcal {M}_{\text {d}}^{{\mathcal {B}}^*}}\) with all three structures. The trend is extremely impressive when comparing against 12 big datasets. Since, each iteration encompasses looping through all the data, these are datasets where each iteration is expensive. It can be seen that \({\mathcal {M}_{\text {w}}^{{\mathcal {B}}^*}}\) achieves a better NLL not only after fifth and tenth iteration, but better even after fiftieth iteration.
Rights and permissions
About this article
Cite this article
Zaidi, N.A., Webb, G.I., Carman, M.J. et al. Efficient parameter learning of Bayesian network classifiers. Mach Learn 106, 1289–1329 (2017). https://doi.org/10.1007/s10994-016-5619-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10994-016-5619-z