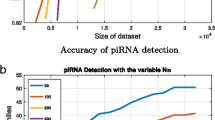
Abstract
Piwi interacting RNA (piRNA) molecules belong to a largest class of small non coding RNA molecules which are originally discovered in animal germline cells and also occur across a variety of human somatic cells. The piRNA molecules play a significant role in many gene functions such as protecting genomic integrity, gene expression regulation and restricting the functions of transposable elements. The identification of piRNA molecules and their function types are significant for cancer cells diagnosis, drug developments and genes stability. A number of traditional machine learning methods have been proposed for identification of piRNAs and their functions. However, these methods are required a considerable amounts of human engineering and expertise to design an accurate identification model. Hence, this paper proposes a two level computational model based on deep neural network (DNN) that automatically extract informative features from RNA sequences using standard learning methods. Moreover, the proposed model employs di-nucleotide auto covariance (DAC) method along with six physiochemical properties to construct a feature vector. The performance of the proposed model has been extensively evaluated through k-fold cross-validation tests. Firstly, the performance of the proposed model is compared with commonly used classifier algorithms using benchmark dataset. Secondly, its performance is compared with the existing state-of- the-art computational models. The experimental results show that the proposed model performed better than the existing predictors with accuracy level 91.81% and 84.52% in the first level and in the second level respectively. The source code along with dataset of the proposed model is freely available at https://github.com/salman-khan-mrd/2L-piRNADNN.






Similar content being viewed by others
References
Acharya UR, Lih S, Hagiwara Y et al (2018) Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med 100:270–278. https://doi.org/10.1016/j.compbiomed.2017.09.017
Alipanahi B, Delong A, Weirauch MT, Frey BJ (2015) Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. https://doi.org/10.1038/nbt.3300
Althaus IW, Chou JJ, Gonzales AJ et al (1993a) Steady-state kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-87201E. J Biol Chem 268(9):6119–6124
Althaus IW, Chou JJ, Gonzales AJ et al (1993b) Kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-88204E. Biochemistry 32:6554–6648
Althaus IW, Gonzales AJ, Chou JJ et al (1993c) The quinoline U-78036 is a potent inhibitor of HIV-1 reverse transcriptase. J Biol Chem 268(20):14875–14880
Althaus IW, Chou JJ, Gonzales AJ et al (1994a) Steady-state kinetic studies with the polysulfonate U-9843, an HIV reverse transcriptase inhibitor. Experientia. https://doi.org/10.1007/bf01992044
Althaus IW, Chou JJ, Gonzales AJ et al (1994b) Kinetic studies with the non-nucleoside human immunodeficiency virus type-1 reverse transcriptase inhibitor U-90152E. Biochem Pharmacol. https://doi.org/10.1016/0006-2952(94)90077-9
Althaus IW, Chou KC, Lemay RJ et al (1996) The benzylthio-pyrimidine U-31,355, a potent inhibitor of HIV-1 reverse transcriptase. Biochem Pharmacol. https://doi.org/10.1016/0006-2952(95)02390-9
Andraos J (2008) Kinetic plasticity and the determination of product ratios for kinetic schemes leading to multiple products without rate laws—new methods based on directed graphs. Can J Chem. https://doi.org/10.1139/v08-020
Aravin A, Gaidatzis D, Pfeffer S et al (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207. https://doi.org/10.1038/nature04916
Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry. W H Free, New York, pp 320–323
Bordes A, Chopra S, Weston J (2014) Question answering with subgraph embeddings. In: Proceedings of the 2014 conference on empirical methods in natural language processing, pp 615–620
Bu D, Yu K, Sun S et al (2012) NONCODE v30: integrative annotation of long noncoding RNAs. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr1175
Carter RE, Forsen S (1981) A new graphical method for driving rate equations for complicated mechanisms. Chem Scr 18:82–86
Chen J, Liu H, Yang J, Chou KC (2007) Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 33:423–428. https://doi.org/10.1007/s00726-006-0485-9
Chen W, Feng PM, Lin H, Chou KC (2013) IRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res 41:1–9. https://doi.org/10.1093/nar/gks1450
Chen W, Lei TY, Jin DC et al (2014) PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal Biochem 456:53–60. https://doi.org/10.1016/j.ab.2014.04.001
Chen W, Tang H, Ye J et al (2016) iRNA-PseU: identifying RNA pseudouridine sites. Mol Ther Nucleic Acids. https://doi.org/10.1038/mtna.2016.37
Chen Y, Li T, Song R et al (2018) Support vector machine classifier for accurate identification of piRNA. Appl Sci. https://doi.org/10.3390/app8112204
Cheng J, Deng H, Xiao B et al (2012) PiR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. https://doi.org/10.1016/j.canlet.2011.10.004
Cheng D, Zhang S, Deng Z et al (2014) kNN algorithm with data-driven k value. In: Luo X, Yu JX, Li Z (eds) Advanced data mining and applications. Springer International Publishing, Cham, pp 499–512
Cheng X, Lin WZ, Xiao X, Chou KC (2019) PLoc-bal-mAnimal: predict subcellular localization of animal proteins by balancing training dataset and PseAAC. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty628
Chou K-C (1989) Graphic rules in steady and non-steady state enzyme kinetics. J Biol Chem 264(20):12074–12079
Chou K-C (1990) Applications of graph theory to enzyme kinetics and protein folding kinetics. Steady and non-steady-state systems. Biophys Chem 35(1):1–24
Chou K-C (2001) Using subsite coupling to predict signal peptides. Protein Eng Des Sel 14:75–79. https://doi.org/10.1093/protein/14.2.75
Chou K-C (2005) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21:10–19. https://doi.org/10.1093/bioinformatics/bth466
Chou K-C (2010) Graphic rule for drug metabolism systems. Curr Drug Metab 11(4):369–378
Chou K-C (2011) Some remarks on protein attribute prediction and pseudo amino acid composition. J Theor Biol 273:236–247
Chou K-C (2015) Impacts of bioinformatics to medicinal chemistry. Med Chem (Los Angeles). https://doi.org/10.2174/1573406411666141229162834
Chou K-C (2017) An unprecedented revolution in medicinal chemistry driven by the progress of biological science. Curr Top Med Chem. https://doi.org/10.2174/1568026617666170414145508
Chou K-C (2019) Advance in predicting subcellular localization of multi-label proteins and its implication for developing multi-target drugs. Curr Med Chem. https://doi.org/10.2174/0929867326666190507082559
Chou K-C, Forsen S (1981) The biological functions of low-frequency phonons: 2 cooperative effects. Chem Scr 1981:126–132
Chou K-C, Forsén S (1980a) Graphical rules for enzyme-catalysed rate laws. Biochem J. https://doi.org/10.1042/bj1870829
Chou K-C, Forsén S (1980b) Diffusion-controlled effects in reversible enzymatic fast reaction systems—critical spherical shell and proximity rate constant. Biophys Chem. https://doi.org/10.1016/0301-4622(80)80002-0
Chou K-C, Shen H-B (2009a) Review: recent advances in developing web-servers for predicting protein attributes. Nat Sci 01:63–92. https://doi.org/10.4236/ns.2009.12011
Chou K-C, Shen H-B (2009b) Review : recent advances in developing web-servers for predicting protein attributes. Nat Sci. https://doi.org/10.4236/ns.2009.12011
Chou K-C, Zhang CT (1995) Prediction of protein structural classes. Crit Rev Biochem Mol Biol 30:275–349. https://doi.org/10.3109/10409239509083488
Chou K-C, Forsen S, Zhou G-Q (1980a) Three schematic rules for deriving apparent rate constants. Chem Scr 16:109–113
Chou K-C, Li TT, Forsén S (1980b) The critical spherical shell in enzymatic fast reaction systems. Biophys Chem. https://doi.org/10.1016/0301-4622(80)80003-2
Chou K-C, Kézdy FJ, Reusser F (1994) Kinetics of processive nucleic acid polymerases and nucleases. Anal. Biochem 221(2):217–230
Chou K-C, Lin W-Z, Xiao X (2011) Wenxiang: a web-server for drawing wenxiang diagrams. Nat Sci. 1:1. https://doi.org/10.4236/ns.2011.310111
Claverie JM (2005) Fewer genes, more noncoding RNA. Science 309:1529–1530
Cox DN, Chao A, Baker J et al (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12:3715–3727. https://doi.org/10.1101/gad.12.23.3715
Dehzangi A, Heffernan R, Sharma A et al (2015) Gram-positive and Gram-negative protein subcellular localization by incorporating evolutionary-based descriptors into Chou’s general PseAAC. J Theor Biol 364:284–294. https://doi.org/10.1016/j.jtbi.2014.09.029
Dong Q, Zhou S, Guan J (2009) A new taxonomy-based protein fold recognition approach based on autocross-covariance transformation. Bioinformatics 25:2655–2662. https://doi.org/10.1093/bioinformatics/btp500
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. https://doi.org/10.1186/1471-2105-8-4
Farabet C, Couprie C, Najman L, LeCun Y (2013) Learning hierarchical features for scene labeling. IEEE Trans Pattern Anal Mach Intell 35:1915–1929. https://doi.org/10.1109/TPAMI.2012.231
Fawagreh K, Gaber MM, Elyan E (2014) Random forests: from early developments to recent advancements. Syst Sci Control Eng 2:602–609. https://doi.org/10.1080/21642583.2014.956265
Fu L, Niu B, Zhu Z et al (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Glorot X, Bengio Y (2010) Understanding the difficulty of training deep feedforward neural networks. PMLR 9:249–256
Grivna ST, Beyret E, Wang Z, Lin H (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20:1709–1714. https://doi.org/10.1101/gad.1434406
Guo Y, Li M, Lu M et al (2006) Predicting G-protein coupled receptors-G-protein coupling specificity based on autocross-covariance transform. Proteins Struct Funct Genet 65:55–60. https://doi.org/10.1002/prot.21097
Guo Y, Yu L, Wen Z, Li M (2008) Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences. Nucleic Acids Res 36:3025–3030. https://doi.org/10.1093/nar/gkn159
Gupta D (2017) Fundamentals of deep learning—activation functions and when to use them. https://www.analyticsvidhya.com/blog/2017/10/fundamentals-deep-learning-activation-functions-when-to-use-them/. Accessed 25 Sep 2018
Harrington S (2017) Gradient descent: high learning rates & divergence
Hashim A, Rizzo F, Marchese G et al (2014) RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 5:9901–9910. https://doi.org/10.18632/oncotarget.2476
Helmstaedter M, Briggman KL, Turaga SC et al (2013) Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500:168
Hinton G, Deng L, Yu D et al (2012) Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process Mag 29:82–97. https://doi.org/10.1109/MSP.2012.2205597
Houwing S, Kamminga LM, Berezikov E et al (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129:69–82. https://doi.org/10.1016/j.cell.2007.03.026
Huang Y, Liu N, Wang JP et al (2012) Regulatory long non-coding RNA and its functions. J Physiol Biochem 68:611–618
Jeong JC, Lin X, Chen X-W (2011) On position-specific scoring matrix for protein function prediction. IEEE/ACM Trans Comput Biol Bioinform 8:308–315. https://doi.org/10.1109/TCBB.2010.93
Jia J, Liu Z, Xiao X et al (2016a) IPPBS-Opt: A sequence-based ensemble classifier for identifying protein-protein binding sites by optimizing imbalanced training datasets. Molecules. https://doi.org/10.3390/molecules21010095
Jia J, Liu Z, Xiao X et al (2016b) ISuc-PseOpt: identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal Biochem 497:48–56. https://doi.org/10.1016/j.ab.2015.12.009
Jiang SP, Liu WM, Fee CH (1979) Graph theory of enzyme kinetics: 1. Steady-state reaction system. Sci Sin 22:341–358
Ju Z, Cao JZ, Gu H (2016) Predicting lysine phosphoglycerylation with fuzzy SVM by incorporating k-spaced amino acid pairs into Chou׳s general PseAAC. J Theor Biol 397:145–150. https://doi.org/10.1016/j.jtbi.2016.02.020
Klattenhoff C, Theurkauf W (2007) Biogenesis and germline functions of piRNAs. Development 135:3–9. https://doi.org/10.1242/dev.006486
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th international conference on neural information processing systems, vol 1. Curran Associates Inc., USA, pp 1097–1105
Kuo-Chen C, Forsen S (2006) Graphical rules of steady-state reaction systems. Can J Chem. https://doi.org/10.1139/v81-107
Lau NC, Seto AG, Kim J et al (2006) Characterization of the piRNA complex from rat testes. Science 313:363–367. https://doi.org/10.1126/science.1130164
Lecun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Leung MKK, Xiong HY, Lee LJ, Frey BJ (2014) Deep learning of the tissue-regulated splicing code. Bioinformatics 30:121–129. https://doi.org/10.1093/bioinformatics/btu277
Li TT, Chou KC (1980) The flow of substrate molecules in fast enzyme catalyzed reaction systems. Chem Scr 16:192–196
Li D, Luo L, Zhang W et al (2016) A genetic algorithm-based weighted ensemble method for predicting transposon-derived piRNAs. BMC Bioinform. https://doi.org/10.1186/s12859-016-1206-3
Lin H, Deng EZ, Ding H et al (2014) IPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res 42:12961–12972. https://doi.org/10.1093/nar/gku1019
Liu B (2017) BioSeq-Analysis: a platform for DNA, RNA, and protein sequence analysis based on machine learning approaches. Brief Bioinform. https://doi.org/10.1093/bib/bbx165
Liu Z, Xiao X, Qiu W-R, Chou K-C (2015) iDNA-Methyl: identifying DNA methylation sites via pseudo trinucleotide composition. Anal Biochem 474:69–77. https://doi.org/10.1016/j.ab.2014.12.009
Liu B, Liu F, Fang L et al (2016) repRNA: a web server for generating various feature vectors of RNA sequences. Mol Genet Genom 291:473–481. https://doi.org/10.1007/s00438-015-1078-7
Liu B, Yang F, Chou KC (2017) 2L-piRNA: a two-layer ensemble classifier for identifying Piwi-interacting RNAs and their function. Mol Ther Nucleic Acids 7:267–277. https://doi.org/10.1016/j.omtn.2017.04.008
Luo L, Li D, Zhang W et al (2016) Accurate prediction of transposon-derived piRNAs by integrating various sequential and physicochemical features. PLoS ONE. https://doi.org/10.1371/journal.pone.0153268
Ma J, Sheridan RP, Liaw A et al (2015) Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model 55:263–274. https://doi.org/10.1021/ci500747n
Mattick JS (2005) The functional genomics of noncoding RNA. Sci (New York, NY) 309:1527–1528. https://doi.org/10.1126/science.1117806
Meenakshisundaram K, Carmen L, Michela B et al (2009) Existence of snoRNA, microRNA, piRNA characteristics in a novel non-coding RNA: x-ncRNA and its biological implication in Homo sapiens. J Bioinform Seq Anal 1:31–40
Mei Y, Clark D, Mao L (2013) Novel dimensions of piRNAs in cancer. Cancer Lett 336:46–52
Miao JH, Miao KH (2018) Cardiotocographic diagnosis of fetal health based on multiclass morphologic pattern predictions using deep learning classification. Int J Adv Comput Sci Appl 9:1–11
Mikolov T, Kombrink S, Burget L, et al (2011) Extensions of recurrent neural network language model. In: 2011 IEEE International conference on acoustics, speech and signal processing (ICASSP), pp 5528–5531
Min S, Lee B, Yoon S (2016) Deep learning in bioinformatics. Brief Bioinform. https://doi.org/10.1093/bib/bbw068
Mondal S, Pai PP (2014) Chou’s pseudo amino acid composition improves sequence-based antifreeze protein prediction. J Theor Biol 356:30–35. https://doi.org/10.1016/j.jtbi.2014.04.006
Moyano M, Stefani G (2015) piRNA involvement in genome stability and human cancer. J Hematol Oncol 8:38. https://doi.org/10.1186/s13045-015-0133-5
Nielsen M (2017) Neural networks and deep learning
Noi PT, Kappas M (2018) Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using sentinel-2 imagery. Sensors (Switzerland). https://doi.org/10.3390/s18010018
Ravi D, Wong C, Deligianni F et al (2017) Deep learning for health informatics. IEEE J Biomed Heal Informatics 21:4–21. https://doi.org/10.1109/JBHI.2016.2636665
Sabooh MF, Iqbal N, Khan M et al (2018) Identifying 5-methylcytosine sites in RNA sequence using composite encoding feature into Chou’s PseKNC. J Theor Biol 452:1–9. https://doi.org/10.1016/j.jtbi.2018.04.037
Sainath TN, Mohamed AR, Kingsbury B, Ramabhadran B (2013) Deep convolutional neural networks for LVCSR. In: 2013 IEEE international conference on acoustics, speech and signal processing. pp 8614–8618
Shen H-B, Song J-N, Chou K-C (2009) Prediction of protein folding rates from primary sequence by fusing multiple sequential features. J Biomed Sci Eng. https://doi.org/10.4236/jbise.2009.23024
Srivastava N, Hinton G, Krizhevsky A et al (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958. https://doi.org/10.1214/12-AOS1000
Tang H, Zou P, Zhang C et al (2016) Identification of apolipoprotein using feature selection technique. Sci Rep 6:1–6. https://doi.org/10.1038/srep30441
Tompson J, Jain A, LeCun Y, Bregler C (2014) Joint training of a convolutional network and a graphical model for human pose estimation. ArXiv e-prints
Tripathi R, Patel S, Kumari V et al (2016) DeepLNC, a long non-coding RNA prediction tool using deep neural network. Netw Model Anal Heal Inform Bioinform 5:21. https://doi.org/10.1007/s13721-016-0129-2
Wang K, Liang C, Liu J et al (2014) Prediction of piRNAs using transposon interaction and a support vector machine. BMC Bioinform. https://doi.org/10.1186/s12859-014-0419-6
Wen Z, Li M, Li Y et al (2007) Delaunay triangulation with partial least squares projection to latent structures: a model for G-protein coupled receptors classification and fast structure recognition. Amino Acids 32:277–283. https://doi.org/10.1007/s00726-006-0341-y
Wikipedia 5-step rules. https://en.wikipedia.org/wiki/5-step_rules. Accessed 25 Jun 2019
Wold S, Jonsson J, Sjörström M et al (1993) DNA and peptide sequences and chemical processes multivariately modelled by principal component analysis and partial least-squares projections to latent structures. Anal Chim Acta 277:239–253. https://doi.org/10.1016/0003-2670(93)80437-P
Xiao X, Cheng X, Chen G et al (2018) pLoc-mGpos: predict subcellular localization of gram-positive bacterial proteins by quasi-balancing training dataset and PseAAC. Genomics. https://doi.org/10.1016/j.ygeno.2018.05.017
Xie C, Yuan J, Li H et al (2014) NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. https://doi.org/10.1093/nar/gkt1222
Xu Y, Ding J, Wu LY, Chou KC (2013) iSNO-PseAAC: predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition. PLoS ONE. https://doi.org/10.1371/journal.pone.0055844
Xu ZC, Wang P, Qiu WR, Xiao X (2017) ISS-PC: identifying splicing sites via physical-chemical properties using deep sparse auto-encoder. Sci Rep 7:1–12. https://doi.org/10.1038/s41598-017-08523-8
Yue S, Li P, Hao P (2003) SVM classification: its contents and challenges. Appl Math J Chinese Univ 18:332–342. https://doi.org/10.1007/s11766-003-0059-5
Zhang Y, Wang X, Kang L (2011) A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics 27:771–776. https://doi.org/10.1093/bioinformatics/btr016
Zhang P, Si X, Skogerbø G et al (2014) PiRBase: a web resource assisting piRNA functional study. Database. https://doi.org/10.1093/database/bau110
Zhou GP (2011) The disposition of the LZCC protein residues in wenxiang diagram provides new insights into the protein-protein interaction mechanism. J Theor Biol. https://doi.org/10.1016/j.jtbi.2011.06.006
Zhou GP, Deng MH (1984) An extension of Chou’s graphic rules for deriving enzyme kinetic equations to systems involving parallel reaction pathways. Biochem J 222(1):169–176
Zhu Z, Albadawy E, Saha A et al (2019) Deep learning for identifying radiogenomic associations in breast cancer. Comput Biol Med 109:85–90. https://doi.org/10.1016/j.compbiomed.2019.04.018
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khan, S., Khan, M., Iqbal, N. et al. A Two-Level Computation Model Based on Deep Learning Algorithm for Identification of piRNA and Their Functions via Chou’s 5-Steps Rule. Int J Pept Res Ther 26, 795–809 (2020). https://doi.org/10.1007/s10989-019-09887-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-019-09887-3