Abstract
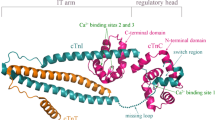
Human cardiac troponin (cTn) is a calcium ion (Ca2+)-sensitive hetero-trimer complex that consists of three subunits: cTnC, cTnI and cTnT. The protein is a primary biomarker and potential target for the diagnosis and therapy of myocardial necrosis in acute coronary syndrome. Previously, a minimal region (mSwt peptide) of cTnI C-terminal switch peptide is identified as a functionally required segment for mediating cTnI interaction with the cTnC N-terminal domain to which Ca2+ binds. Here, we attempt to investigate the Ca2+-dependent effect on the inter-subunit interaction between cTnC and cTnI at molecular level. The structure of Ca2+-bound and Ca2+-free cTnC N-terminal domains as well as their complexes with mSwt peptide are stripped from the crystal structure of full-length cTn complex, which are then subjected to 600-ns molecular dynamics simulations for conformational equilibrium and post energetics analysis. It is revealed that Ca2+ can effectively stabilize the native conformation of cTnC N-terminal domain and the tight binding mode of mSwt peptide to the domain; lack of Ca2+ would cause a considerable unfolding phenomenon in the domain, particularly in helices H1 and H5, and largely shift the interaction manner between the domain and peptide. In addition, there is also a significant difference between the binding curves of mSwt peptide to Ca2+-bound and Ca2+-free domains along the dynamics trajectory; the former monotonically increases over the whole simulations, whereas the latter exhibits a unimodal profile. Binding analysis also observes a significant concentration-dependent effect of Ca2+ on the domain–peptide affinity, that is, just a little of Ca2+ can improve the affinity remarkably. Overall, it is demonstrated that Ca2+ plays a crucial role in domain–peptide interaction and Ca2+ should bind very efficiently to the domain.






Similar content being viewed by others
References
Althaus IW, Gonzales AJ, Chou JJ, Diebel MR, Kezdy FJ, Romero DL, Aristoff PA, Tarpley WG, Reusser F (1993a) The quinoline U-78036 is a potent inhibitor of HIV-1 reverse transcriptase. J Biol Chem 268:14875–14880
Althaus IW, Chou JJ, Gonzales AJ, Diebel MR, Kezdy FJ, Romero DL, Aristoff PA, Tarpley WG, Reusser F (1993b) Steady-state kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-87201E. J Biol Chem 268:6119–6124
Althaus IW, Chou JJ, Gonzales AJ, Diebel MR, Kezdy FJ, Romero DL, Aristoff PA, Tarpley WG, Reusser F (1993c) Kinetic studies with the nonnucleoside HIV-1 reverse transcriptase inhibitor U-88204E. Biochemistry 32:6548–6554
Bai Z, Hou S, Zhang S, Li Z, Zhou P (2017) Targeting self-binding peptides as a novel strategy to regulate protein activity and function: a case study on the proto-oncogene tyrosine protein kinase c-Src. J Chem Inf Model 57:835–845
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 2:235–242
Cheng X, Xiao X (2017) pLoc-mPlant: predict subcellular localization of multi-location plant proteins via incorporating the optimal GO information into general PseAAC. Mol BioSyst 13:1722–1727
Chou KC (1989) Graphic rules in steady and non-steady enzyme kinetics. J Biol Chem 264:12074–12079
Chou KC (1990) Review: applications of graph theory to enzyme kinetics and protein folding kinetics. Steady and non-steady state systems. Biophys Chem 35:1–24
Chou KC (2001) Prediction of protein cellular attributes using pseudo amino acid composition. Proteins 43:246–255
Chou KC (2005a) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21:10–19
Chou KC (2005b) Coupling interaction between thromboxane A2 receptor and alpha-13 subunit of guanine nucleotide-binding protein. J Proteome Res 4:1681–1686
Chou KC (2011) Some remarks on protein attribute prediction and pseudo amino acid composition. J Theor Biol 273:236–247
Chou KC (2015) Impacts of bioinformatics to medicinal chemistry. Med Chem 11:218–234
Chou KC (2017) An unprecedented revolution in medicinal chemistry driven by the progress of biological science. Curr Top Med Chem 17:2337–2358
Chou KC, Chen NY (1977) The biological functions of low-frequency phonons. Sci Sinica 20:447–457
Chou KC, Forsen S (1980) Graphical rules for enzyme-catalyzed rate laws. Biochem J 187:829–835
Chou KC, Forsen S (1981) Graphical rules of steady-state reaction systems. Can J Chem 59:737–755
Chou KC, Jiang SP, Liu WM, Fee CH (1979) Graph theory of enzyme kinetics: 1. Steady-state reaction system. Sci Sin 22:341–358
Chou KC, Forsen S, Zhou GQ (1980) Three schematic rules for deriving apparent rate constants. Chem Scr 16:109–113
Chou KC, Carter RE, Forsen S (1981) A new graphical method for deriving rate equations for complicated mechanisms. Chem Scr 18:82–86
Chou KC, Zhang CT, Maggiora GM (1994) Solitary wave dynamics as a mechanism for explaining the internal motion during microtubule growth. Biopolymers 34:143–153
Chou KC, Lin WZ, Xiao X (2011) Wenxiang: a web-server for drawing wenxiang diagrams. Nat Sci 3:862–865
Darden T, York D, Pedersen L (1983) Particale mesh Ewald and N.log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Davis JP, Tikunova SB (2008) Ca2+ exchange with troponin C and cardiac muscle dynamics. Cardiovasc Res 77:619–626
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM, Zhang W (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Haikala H, Linden IB (1995) Mechanisms of action of calcium-sensitizing drugs. J Cardiovasc Pharmacol 26:S10–S19
Huang RB, Du QS, Wang CH (2008) An in-depth analysis of the biological functional studies based on the NMR M2 channel structure of influenza A virus. Biochem Biophys Res Commun 377:1243–1247
Hussain W, Khan YD, Rasool N, Khan SA (2019a) SPalmitoylC-PseAAC: a sequence-based model developed via Chou’s 5-steps rule and general PseAAC for identifying S-palmitoylation sites in proteins. Anal Biochem 568:14–23
Hussain W, Khan YD, Rasool N, Khan SA (2019b) SPrenylC-PseAAC: a sequence-based model developed via Chou’s 5-steps rule and general PseAAC for identifying S-prenylation sites in proteins. J Theor Biol 468:1–11
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Phys Chem 79:926–935
Li MX, Robertson IM, Sykes BD (2008) Interaction of cardiac troponin with cardiotonic drugs: a structural perspective. Biochem Biophys Res Commun 369:88–99
Li Z, Yan F, Miao Q, Meng Y, Wen L, Jiang Q, Zhou P (2019a) Self-binding peptides: binding-upon-folding versus folding-upon-binding. J Theor Biol 469:25–34
Li Z, Miao Q, Yan F, Meng Y, Zhou P (2019b) Machine learning in quantitative protein-peptide affinity prediction: implications for therapeutic peptide design. Curr Drug Metab 20:170–176
Liu B, Liu F, Wang X, Chen J, Fang L (2015) Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Res 43:W65–W71
Luo H, Du T, Zhou P, Yang L, Mei H, Ng H, Zhang W, Shu M, Tong W, Shi L, Mendrick DL, Hong H (2015) Molecular docking to identify associations between drugs and class I human leukocyte antigens for predicting idiosyncratic drug reactions. Comb Chem High Throughput Screen 18:296–304
Qiu WR, Jiang SY, Xu ZC, Xiao X (2017) iRNAm 5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget 8:41178–41188
Ren Y, Chen X, Feng M, Wang Q, Zhou P (2011) Gaussian process: a promising approach for the modeling and prediction of peptide binding affinity to MHC proteins. Protein Pept Lett 18:670–678
Ryckaert JP, Ciccotti G, Berendsen HJC (1997) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Saíz-Urra L, Cabrera MA, Froeyen M (2011) Exploring the conformational changes of the ATP binding site of gyrase B from Escherichia coli complexed with different established inhibitors by using molecular dynamics simulation: protein-ligand interactions in the light of the alanine scanning and free energy decomposition methods. J Mol Graph Model 29:726–739
Schnell JR, Chou JJ (2008) Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451:591–595
Sharma S, Jackson PG, Makan J (2004) Cardiac troponins. J Clin Pathol 57:1025–1026
Takeda S, Yamashita A, Maeda K, Maéda Y (2003) Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature 424:35–41
Tian F, Lv Y, Zhou P, Yang L (2011) Characterization of PDZ domain-peptide interactions using an integrated protocol of QM/MM, PB/SA, and CFEA analyses. J Comput Aided Mol Des 25:947–958
Tian F, Tan R, Guo T, Zhou P, Yang L (2013) Fast and reliable prediction of domain-peptide binding affinity using coarse-grained structure models. Biosystems 113:40–49
Tian F, Yang C, Wang C, Guo T, Zhou P (2014) Mutatomics analysis of the systematic thermostability profile of Bacillus subtilis lipase A. J Mol Model 20:2257
Wang SQ, Du QS (2007) Study of drug resistance of chicken influenza A virus (H5N1) from homology-modeled 3D structures of neuraminidases. Biochem Biophys Res Commun 354:634–640
White SP, Cohen C, Phillips GN (1987) Structure of co-crystals of tropomyosin and troponin. Nature 325:826–828
Xiao X, Min JL, Lin WZ, Liu Z, Cheng X (2015) iDrug-Target: predicting the interactions between drug compounds and target proteins in cellular networking via the benchmark dataset optimization approach. J Biomol Struct Dyn 33:2221–2233
Xiao X, Cheng X, Chen G, Mao Q (2018) pLoc_bal-mGpos: predict subcellular localization of Gram-positive bacterial proteins by quasi-balancing training dataset and PseAAC. Genomics 26:S0888–S7543
Xu Y, Huang R, Gu J, Jiang W (2017) Derivation of inhibitory peptides to target the cardiac troponin C-I interaction as potential therapeutics for heart failure. Int J Pept Res Ther 23:387–392
Yang Y, Liu H, Yao X (2012) Understanding the molecular basis of MK2-p38α signaling complex assembly: insights into protein-protein interaction by molecular dynamics and free energy studies. Mol BioSyst 8:2106–2118
Yang C, Wang C, Zhang S, Huang J, Zhou P (2015a) Structural and energetic insights into the intermolecular interaction among human leukocyte antigens, clinical hypersensitive drugs and antigenic peptides. Mol Simul 41:741–751
Yang C, Zhang S, He P, Wang C, Huang J, Zhou P (2015b) Self-binding peptides: folding or binding. J Chem Inf Model 55:329–342
Yang C, Zhang S, Bai Z, Hou S, Wu D, Huang J, Zhou P (2016) A two-step binding mechanism for the self-binding peptide recognition of target domains. Mol BioSyst 12:1201–1213
Yu H, Zhou P, Deng M, Shang Z (2014) Indirect readout in protein-peptide recognition: a different story from classical biomolecular recognition. J Chem Inf Model 54:2022–2032
Zhou GP (2011) The disposition of the LZCC protein residues in Wenxiang diagram provides new insights into the protein-protein interaction mechanism. J Theor Biol 284:142–148
Zhou GP, Deng MH (1984) An extension of Chou’s graphic rules for deriving enzyme kinetic equations to systems involving parallel reaction pathways. Biochem J 222:169–176
Zhou P, Wang C, Tian F, Ren Y, Yang C, Huang J (2013a) Biomacromolecular quantitative structure-activity relationship (BioQSAR): a proof-of-concept study on the modeling, prediction and interpretation of protein-protein binding affinity. J Comput Aided Mol Des 27:67–78
Zhou P, Yang C, Ren Y, Wang C, Tian F (2013b) What are the ideal properties for functional food peptides with antihypertensive effect? A computational peptidology approach. Food Chem 141:2967–2973
Zhou P, Zhang S, Wang Y, Yang C, Huang J (2016) Structural modeling of HLA-B*1502 peptide carbamazepine T-cell receptor complex architecture: implication for the molecular mechanism of carbamazepine-induced Stevens-Johnson syndrome toxic epidermal necrolysis. J Biomol Struct Dyn 34:1806–1817
Zhou P, Hou S, Bai Z, Li Z, Wang H, Chen Z, Meng Y (2018) Disrupting the intramolecular interaction between proto-oncogene c-Src SH3 domain and its self-binding peptide PPII with rationally designed peptide ligands. Artif Cells Nanomed Biotechnol 46:1122–1131
Zhou P, Miao Q, Yan F, Li Z, Jiang Q, Wen L, Meng Y (2019) Is protein context responsible for peptide-mediated interactions? Mol Omics. https://doi.org/10.1039/c9mo00041k
Acknowledgements
The authors are grateful to Mr. Yingjia Xu for valuable discussions and software helps. This work was supported by the AJHJU foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yu, W., Lv, Y., Ding, Y. et al. Studying Calcium Ion-Dependent Effect on the Inter-subunit Interaction Between the cTnC N-terminal Domain and cTnI C-terminal Switch Peptide of Human Cardiac Troponin via Chou’s 5-Steps Rule. Int J Pept Res Ther 26, 675–683 (2020). https://doi.org/10.1007/s10989-019-09875-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-019-09875-7