Abstract
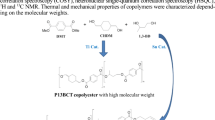
In order to understand the relationship between enzymatic degradation and the structure of PBS-based copolyesters PBS-co-DEGS and PBS-co-BDGA modified with monomers (diethylene glycol and diglycolic acid) were synthesized and the enzymatic degradation property was studied. Pseudomonas cepacia lipase was selected as the catalyst, while the chloroform was used as solvent. The results indicated that both PBS-co-DEGS and PBS-co-BDGA can be catalytically degraded by PC lipase but they were different. The hydrolysis of PBS-co-BDGA improved greatly and the degree of hydrolysis of PBS-co-BDGA was almost consistent with the degree of degradation due to the introduction of DGA. Similarly, thermal property changes were also observed with a decrease of the decomposition temperature of 5 and 50% sample in most cases. The enzymatic degradation of PBS-based copolymers produced not only linear segments, but also cyclic oligomers. Furthermore, PBS-co-BDGA generated more oligomers than PBS-co-DEGS. According to the results of molecular docking, the free energy of binding between PCL and the substrate in chloroform was in the order BDGAB > DEGSDEG > BSB. That is, the docking of the substrate containing BDGA in the active site of PCL was more stable than any other ones.














Similar content being viewed by others
Abbreviations
- DEG:
-
Diethylene glycol
- DGA:
-
Diglycolic acid
- SA:
-
Succinic acid
- BDO:
-
1, 4-butanediol
- BDGA:
-
Butylene diglycolic acid
- DEGS:
-
Diethylene glycol succinate
- BSB:
-
Butylene succinate butylene
- BDGAB:
-
Butylene diglycolic acid butylene
- DEGSDEG:
-
Diethylene glycol succinate diethylene
- PBS:
-
Poly(butylene succinate)
- PBS-co-DEGS:
-
Poly (butylene succinate-co-diethylene glycol succinate)
- PBS-co-BDGA:
-
Poly poly(butylene succinate-co-butanediol diethylene glyco alkyd
- P(BS-co-5%DEGS):
-
Poly (butylene succinate-co-5%diethylene glycol succinate)
- P(BS-co-5%BDGA):
-
Poly (butylene succinate-co-butylene 5%diglycolic acid)
- CHCl3 :
-
Chloroform
- PC lipase:
-
Pseudomonas cepacia lipase
- MALDI-TOF-MS:
-
Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry
- GPC:
-
Gel Permeation Chromatography
References
Azapagic A, Emsley A, Hamerton I (2003) Polymers: the environment and sustainable development. John Wiley & Sons
Sivan A (2011) New perspectives in plastic biodegradation. Curr Opin Biotechnol 22:422–426
Imre B, Pukánszky B (2013) Compatibilization in bio-based and biodegra- dable polymer blends. Eur Polym J 49:1215–1233
Gross RA, Kalra B (2002) Biodegradable polymers for the environment. Science 297:803–807
Mahalik JP, Madras G (2006) Enzymatic degradation of poly (D,L-lactide) and its, blends with poly(vinyl acetate). J Appl Polym Sci 101:675–680
Tsuji H, Miyauchi S (2001) Poly (l-lactide). 7. Enzymatic hydrolysis of free and restricted amorphous regions in poly(l-lactide) films with different crystallinities and a fixed crystalline thickness. Polymer 42:4463–4467
Kurokawa K, Yamashita K, Doi Y, Abe H (2006) Surface properties and enzymatic degradation of end-capped poly(L-lactide). Polym Degrad Stabil 91:1300–1310
Qin J, Zhang M, Zhang C, Li C, Zhang Y, Song J, Asif Javed HM, Qiu J (2016) New insight into the difference of PC lipase-catalyzed degradation on poly (butylene succinate)-based copolymers from molecular levels. RSC Adv 6:17896–17905
Kobayashi S, Uyama H, Takamoto T (2000) Lipase-catalyzed degradation of polyesters in organic solvents. A new methodology of polymer recycling using enzyme as catalys. Biomacromolecules 1:3–5
Rapaport DC (2004) The art of molecular dynamics simulation. Cambridge University Press
Sun X, Ågren H, Tu Y (2014) Microsecond molecular dynamics simulations provide insight into the allosteric mechanism of the Gs protein uncoupling from the β2 adrenergic receptor. J Phys Chem B 118:14737–14744
Sun X, Ågren H, Tu Y (2014) Functional water molecules in rhodopsin activation. J Phys Chem 118:10863–10873
Qin J, Luo W, Li M, Chen P, Wang S, Ren S, Han D, Xiao M, Meng Y (2017) A novel multiblock copolymer of CO2-based PPC-mb-PBS: from simulation to experiment. ACS Sustain Chem Eng 5:5922–5930
Zhou H, Qu Y, Kong C, Wang J, Zhang X, Ma Q, Zhou J (2013) The key role of a non-active-site residue Met148 on the catalytic efficiency of meta-cleavage product hydrolase BphD. Appl Microbiol Biotechnol 97:10399–10411
Krieger E, Koraimann G, Vriend G (2002) Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins 47:393–402
Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57:678–683
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J Kollman P(2003)a point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Luic M, Šytefanic Z, Ceilinger I, Hodoscek M, Janežic D, Lenac T, Ašler IL, Šyepac D, Tomic S (2008) Combined X-ray diffraction and QM/MM study of the Burkholderia cepacia lipase-catalyzed secondary alcohol esterification. J Phys Chem B 112:4876–4883
Ding M, Zhang M, Yang J, Qiu J (2012) Study on the enzymatic degradation of PBS and its alcohol acid modified copolymer. Biodegradation 23:127–132
Zhang M, Ding M, Zhang T, Yang J (2010) Effect of solvent on the enzyme catalysis biodegradation of PBS with high molecular weight and its modified copolymer. Chem J Chin Univ 3:612–615
Soulis S, Triantou D, Weidner S, Falkenhagen J, Simitzis J (2012) Structural analysis of biodegradable low-molecular mass copolyesters based on glycolic acid, adipic acid and 1,4 butanediol and correlation with their hydrolytic degradation. Polym Degrad Stab 97:2091–2103
Acknowledgements
Financial support for this work was provided by the Research Fund of the People’s Government of Shaanxi Province of China (2016CG-10), Shaanxi University of Science and Technology (BJ15-21), Beijing Key Laboratory of Quality Evaluation Technology for Hygiene and Safety of Plastics, Beijing Technology and Business University (BS201703) and Shaanxi Educational Committee (17JK0111).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Li, Ct., Zhang, M., Weng, Yx. et al. Influence of ether bond on degradation property of PBS-based copolymers at molecular level using molecular simulations. J Polym Res 25, 161 (2018). https://doi.org/10.1007/s10965-018-1560-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10965-018-1560-0