Abstract
Lakes cover large parts of the climatically sensitive Arctic landscape and respond rapidly to environmental change. Arctic lakes have different origins and include the predominant thermokarst lakes, which are small, young and highly dynamic, as well as large, old and stable glacial lakes. Freshwater diatoms dominate the primary producer community in these lakes and can be used to detect biotic responses to climate and environmental change. We used specific diatom metabarcoding on sedimentary DNA, combined with next-generation sequencing and diatom morphology, to assess diatom diversity in five glacial and 15 thermokarst lakes within the easternmost expanse of the Siberian treeline ecotone in Chukotka, Russia. We obtained 163 verified diatom sequence types and identified 176 diatom species morphologically. Although there were large differences in taxonomic assignment using the two approaches, they showed similar high abundances and diversity of Fragilariceae and Aulacoseiraceae. In particular, the genetic approach detected hidden within-lake variations of fragilarioids in glacial lakes and dominance of centric Aulacoseira species, whereas Lindavia ocellata was predominant using morphology. In thermokarst lakes, sequence types and valve counts also detected high diversity of Fragilariaceae, which followed the vegetation gradient along the treeline. Ordination analyses of the genetic data from glacial and thermokarst lakes suggest that concentrations of sulfate (SO42−), an indicator of the activity of sulfate-reducing microbes under anoxic conditions, and bicarbonate (HCO3−), which relates to surrounding vegetation, have a significant influence on diatom community composition. For thermokarst lakes, we also identified lake depth as an important variable, but SO42− best explains diatom diversity derived from genetic data, whereas HCO3− best explains the data from valve counts. Higher diatom diversity was detected in glacial lakes, most likely related to greater lake age and different edaphic settings, which gave rise to diversification and endemism. In contrast, small, dynamic thermokarst lakes are inhabited by stress-tolerant fragilarioids and are related to different vegetation types along the treeline ecotone. Our study demonstrated that genetic investigations of lake sediments can be used to interpret climate and environmental responses of diatoms. It also showed how lake type affects diatom diversity, and that such genetic analyses can be used to track diatom community changes under ongoing warming in the Arctic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arctic and high-elevation permafrost regions have recently warmed faster than areas elsewhere on Earth (Huang et al. 2017; Biskaborn et al. 2019a). Thus, dramatic socio-economic and ecological consequences are expected (AMAP 2017). In particular, Arctic lakes represent early-warning systems of environmental change in these areas, as they dominate the landscape and respond rapidly to climate perturbations and ecological changes in lake catchments. Lakes in northern Siberia have multiple origins that, among others, include glacial and thermokarst processes. It is widely believed that north-eastern Siberia was largely ice-free during the Last Glacial Maximum (LGM) (Svendsen et al. 2004) and that glaciation of continental areas in north-eastern Siberia likely occurred in earlier glacial periods during the Quaternary, which led to the formation of glacial lakes as a result of retreating ice sheets in mountain areas (Gualtieri et al. 2000). Growth of ice sheets in northern Russia occurred mainly during the early and middle Weichselian (MIS 4–3) and it is believed that most of Siberia has been ice-free since 50,000 years ago (Svendsen et al. 2004). Therefore, glacially formed lakes in Siberian mountain regions represent accumulation basins that have a long history of ecosystem development, with respect to both the water-filled basins themselves and the surrounding catchments.
Glacial lakes in Russia are often characterised by deep and stratified waters, which during winter are covered by a thick ice layer (up to several metres), but do not freeze to the ground (Biskaborn et al. 2019b). These large and old lakes possess a wide range of established, in-lake habitats and heterogenous catchment areas that impact intra-lake biodiversity patterns, as has been shown in sub-arctic Lake Bolshoe Toko (Biskaborn et al. 2019b; Stoof-Leichsenring et al. 2020). Moreover, studies on large lakes uncovered the presence of endemic species (Cvetkoska et al. 2018; Genkal and Yarushina 2018) and are hotspots of biodiversity (Cvetkoska et al. 2018).
The Arctic landscape is commonly dotted with a wide range of different-size lakes and ponds (Grosse et al. 2013). Among these lakes, thermokarst lakes are the predominant type in Arctic permafrost regions (Bouchard et al. 2016) and comprise around 98% of the Arctic lakes (Wik et al. 2016). They started to form at the transition from the Pleistocene cold period to the warm Holocene, with formation peaking during the Holocene Thermal Maximum as a consequence of degradation of ice-rich permafrost (Schleusner et al. 2015). Since they originated from ground subsidence as a result of ground-ice thawing, they are mostly characterised by shallow water depth and small area, depending on the initial ground-ice distribution and the sediment supply (Subetto et al. 2017). Many shallow thermokarst lakes in the continental climate of Siberia freeze to the bottom during the long winter (Pestryakova et al. 2012), and experience complete water mixing after ice break-up, with very short phases of thermal stratification in the ice-free period (Boike et al. 2015). Because of climate warming and permafrost degradation, they have a tendency to undergo expansion and eventual drainage (Biskaborn et al. 2013; Lenz et al. 2016). Moreover, the hydrochemical variability of thermokarst lakes depends on land-cover changes in the catchment (Bouchard et al. 2016). In general, highly dynamic thermokarst lakes host diverse biota that can withstand disturbances and adapt easily to new environmental conditions (Biskaborn et al. 2012).
In order to analyse the link between biodiversity changes and lake type, it is important to account for the differences in limno-ecological properties, which are reflected by the biological remains in sediments that record the recent ecosystem status. Predominant components of lake sediments, including glacial and thermokarst lakes, are living and dead diatoms. Diatoms are environmentally sensitive, photosynthetic microalgae and are established biological indicators, frequently used to evaluate and monitor modern water quality (Krammer and Lange-Bertalot 1986–1991), and reconstruct palaeoenvironmental conditions (Smol and Stoermer 2010). Temporal succession of diatoms in lake sediment records can provide valuable information on the post-glacial environmental development of a lake and its catchment (Laing and Smol 2000; Biskaborn et al. 2012, 2013). Quantitative estimates of past limnological variables are necessary, and training datasets comprised of diatom assemblages from surface sediments, together with modern limnological observations, are required to develop transfer functions that can be used to infer past environmental changes responsible for diatom community changes (Pestryakova et al. 2012, 2018; Herzschuh et al. 2013). The identification of diatoms is traditionally performed by morphological inspection of silicified diatom frustules, using a light microscope. In recent decades, metabarcoding approaches on environmental DNA from lake sediment samples have been applied successfully to assess diatom community composition (Epp et al. 2015; Dulias et al. 2017). Recent studies confirmed the strong relationship between morphologically and genetically determined diatom community composition in sediments, by comparing metabarcoding approaches and traditional valve counts in lakes and streams (Zimmermann et al. 2015; Dulias et al. 2017). DNA metabarcoding approaches enable the detection of morphologically hidden diversity, depending on the resolution of the applied genetic marker and the completeness of the reference database (Kermarrec et al. 2013; Guardiola et al. 2015; Zimmermann et al. 2015). A few studies, based on morphological diatom analyses in glacially formed Lake Bolshoe Toko, Siberia, documented the persistence of endemic diatom taxa (Genkal and Yarushina 2018; Biskaborn et al. 2019b). The metabarcoding studies on Siberian thermokarst lakes indicate hidden and cryptic diversity in diatom communities, and revealed intra-specific variation in small fragilariod taxa, and their spatial and temporal distribution patterns (Stoof-Leichsenring et al. 2014, 2015). These distribution patterns were related to vegetation changes in the lake catchments on a transect across the Siberian treeline ecotone (Stoof-Leichsenring et al. 2015), which are known to affect ion concentrations in lake waters (Rühland et al. 2003; Biskaborn et al. 2012; Herzschuh et al. 2013).
Our study aimed to gain insights into differences in diatom communities from glacial and thermokarst lakes sampled along the easternmost extent of the Siberian treeline ecotone in far north-eastern Chukotka, Russian Arctic. We applied metabarcoding, combined with next-generation sequencing and classical morphological investigations of diatoms from surface sediments to: (1) reveal the taxonomic resolution and diatom community patterns obtained from genetic and morphological diatom identification, and (2) identify the environmental drivers of diatom communities in the main lake types (glacial and thermokarst) across the treeline ecotone.
Materials and methods
Sampling and collection of environmental data
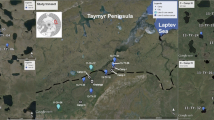
In summer 2016, sediments and water samples were collected from 21 lakes in Chukotka, north-eastern Siberia, Russia (Fig. 1a–e) from four areas traversing a southwest-northeast transect across the Siberian forest-tundra ecotone. Large, glacially formed lakes (mean area 10 km2, mean maximum depth 25.5 m) were sampled at a number of within-lake locations (n = 20 samples) and 16 small thermokarst lakes (mean area 0.1 km2, mean maximum depth 7.5 m) were sampled at one site. At each lake, we recorded geographic position using a hand-held Garmin GPS device, observed vegetation, and physical (lake area, lake depth, Secchi depth) and chemical variables (pH, conductivity) (Electronic Supplementary Material [ESM] Table S1). Water depth at each sampling locality was measured using a hand-held ECHOTEST sounder. Bathymetric maps (Fig. 1) were estimated from good coverage of water-depth measurements taken during sub-bottom profiling in summer 2018 for Lakes Ilirney and Rauchuagytgyn. A simple water-depth map for Lake Nutenvut was estimated using a single profile of echo-sound data from north to south, obtained during fieldwork in 2016, because no seismic measurements were performed on this lake. Surface sediments were collected with a bottom sampler acc. to Lenz and the uppermost cm of surface sediment was transferred to sterile bottles using sterile spoons, whilst wearing gloves. Water samples were collected with a water sampler and stored in labelled sample tubes for subsequent water chemistry analysis. Sediment and water samples were transported under dark and cool conditions and were subsequently analysed for DNA and water chemistry in the laboratories of the Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research (AWI) in Potsdam, Germany, and for diatom remains at the North-Eastern Federal University of Yakutsk, Russia.
a Locations of the four lake sites in Eastern Russia from which surface-sediment samples were collected. b Field site 16-KP-01: samples were collected from glacial Lake Ilirney (ILI, 9 samples) and from four adjacent thermokarst lakes. c Field site 16-KP-02: samples were collected from four thermokarst lakes. d Field site 16-KP-04: four samples were collected from glacial Lake Rauchuagytgyn (RAU) and from three adjacent thermokarst lakes. e Field site 16-KP-03: five samples were collected from glacial Lake Nutenvut (NUT), two samples from smaller glacial lakes and five samples from adjacent thermokarst lakes. The Normalised Difference Vegetation Index (NDVI) utilises near infrared and red bandwidths to assess chlorophyll content and represents photosynthetic capacity of the vegetation canopy. NDVI ranges from 0 (no vegetation) to 1 (dense healthy vegetation). Bathymetric maps were made using either a hand-held echo sounder in summer 2016 (Lake Nutenvut) or by parametric sub-bottom profiling in summer 2018 (Lakes Ilirney and Rauchuagytgyn)
Diatom genetic assessment
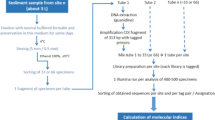
Thirty-six lake sediment samples were prepared for genetic analysis. About 5 g of each surface sediment sample were transferred with a sterile spatula into sterile Falcon tubes and stored at − 20 °C for further processing. DNA isolation was performed using a DNeasy PowerMax Soil Kit (Qiagen, Germany) and was processed in an isolation laboratory with an ultraviolet hood used exclusively for environmental DNA extraction in an isolation laboratory separated from the Post-PCR area. Fifteen mL of bead solution, 1.2 mL C1 buffer, 400 µl of 2 mg/L proteinase K (VWR International) and 100 µL of 5 M dithiothreitol were prepared in a bead tube for each sample. 1.5 mL of the buffer solution mixture was added to each sediment sample tube to liquidize the sticky sediment sample. After that, the sample-buffer mixture was then poured back into the corresponding bead tube and vortexed for 10 min at maximum speed and incubated overnight at 56 °C in a rocking shaker. The following extraction steps were carried out according to the kit manufacturer’s instructions. In the final elution step, 1.6 mL elution buffer was used and the incubation time was extended to 10 min. Isolated DNA was stored at − 20 °C. In total, five extraction batches were processed. To check for chemical contamination, one extraction blank was included for each extraction batch. PCR protocols were set up with diatom-specific primers that amplify a short DNA fragment (76 bp without primer sequence) of the rbcL gene (Stoof-Leichsenring et al. 2012). To distinguish the samples after sequencing, both forward and reverse primers were modified by adding eight unique, randomised nucleotide tags on the 5′ end and three additional unidentified bases (NNN), to improve cluster detection on Illumina sequencing platforms (De Barba et al. 2014). The reactions were prepared with the following reagents: Primers (forward: 5′ NNN(8 bp tag)AACAGGTGAAGTTAAAGGTTCATAYTT 3′, reverse: 5′ NNN(8 bp tag)TGTAACCCATAACTAAATCGATCAT 3′) each primer has an end concentration of 0.5 µM, 10 × Platinum® Taq DNA Polymerase High Fidelity PCR buffer (Invitrogen, USA), 2.5 mM deoxyribonucleotide triphosphate, 0.5 mg Bovine Serum Albumin, 50 mM MgSO4 (Invitrogen, USA), 1.25 U Platinum® Taq High Fidelity DNA Polymerase (Invitrogen, USA) and 2 µL of DNA template. PCR set-ups were performed under a dedicated ultraviolet working station separated from the Post-PCR area. PCRs were run in the Post-PCR area and were performed in a Biometra thermo cycler (Jena Analytik, Germany) with the conditions of initial denaturation at 94 °C for 5 min, followed by 50 cycles at 94, 49 and 68 °C each for 30 s and a final extension at 72 °C for 10 min. The PCRs were performed in three replicates using different primer tag combinations. Twelve PCR reactions were performed and nine to eleven sediment samples were included in each reaction. PCR negative control (NTC) and the extraction blank were run alongside each reaction. In total, there were 141 PCR products, including replicates, extraction blanks and NTCs. The expected amplifications were assessed by 2% agarose (Carl Roth GmbH and Co. KG, Germany) gel electrophoresis.
After PCR and gel evaluation, all PCR products were purified using the MinElute PCR Purification Kit (Qiagen, Germany), following the kit manufacturer’s instructions. The purified PCR products were eluted to a final volume of 20 μL. For DNA concentration measurement, 1 μL of DNA was quantified using the Qubit® dsDNA BR Assay Kit (Invitrogen, USA). In each sample, a certain volume was calculated based on the measured concentration, to ensure 60 ng DNA for sequencing. All samples were then pooled equimolarly to a final concentration of approximately 1000 ng in 30 μL. NTCs and extraction blanks were adjusted to a volume of 10 μL and were added to the pool. Library preparations, according to the specifications of the MetaFast protocol (developed by Fasteris), and parallel high-throughput paired-end (2 × 125 bp) amplicon sequencing, were performed on the Illumina HiSeq 2500 platform (Illumina Inc.) conducted by Fasteris SA sequencing service (Switzerland).
Raw sequence processing and taxonomic assignment
Sequence data were processed using the OBITools package (Boyer et al. 2016). The raw sequencing data, consisting of two single fastq data files, which were first assembled to a single file using the algorithm illuminapairedend, and sequences having a low alignment quality score (threshold set at 40) were filtered out. The retained sequences were assigned to the samples according to their corresponding tag combinations using ngsfilter, by matching 100% with tags. As the same DNA molecule can be sequenced several times, identical sequences were then summarised using the obiuniq command. To de-noise the data from rare reads that are possibly PCR and/or sequencing errors, obigrep was used to discard sequences with less than 10 read counts. As a final de-noising step, obiclean was used to exclude further sequence variants, probably attributable to PCR and/or sequencing errors, by classifying the sequences into head, internal and singleton based on the count and sequence similarity within one sample.
The reference database used for taxonomic assignments was created with the ecoPCR program using the primer pairs mentioned above to simulate a PCR (Boyer et al. 2016). This in silico PCR (Ficetola et al. 2010) was performed on the EMBL Nucleotide Database (Release 133, October 2017) with three mismatches between primers and target sequences. The formatted ecoPCR output was then filtered by obigrep to ensure the taxonomic resolutions are at the species, genus, and family levels. Obiuniq was used to further de-replicate the redundant sequences. The sequences went through obigrep again to ensure a taxid at the family level. Finally, the reference database was prepared using obiconvert to format the filtered database into an ecoPCR-compatible file.
The processed sequences were assigned using ecotag by searching for possible matches based on sequence similarity with the reference library. The threshold of the similarity was set to 0.90 to allow 10% misidentifications of the sequences to the reference library entries. We chose this threshold because of the incompleteness of the reference database with respect to polar continental diatoms. With a low threshold, we prevent sequence types that have no exact reference in the database from being excluded from our dataset. Precautions were taken to further filter the sequences to have reliable taxonomic assignment of the sequences, but not to over-estimate the community diversity. As the targeted diatom region was expected to be 76 bp, only assigned sequences that contain the exact length of 76 bp were kept. Sequences that did not belong to the phylum Bacillariophyta were also excluded. Rare sequences occurring with less than 10 read counts across the dataset were replaced with 0, as probable artefacts. Sequence types that occurred less than three times in all the PCR batches, including PCR replicates, were also discarded, as were diatom sequence types with a count less than 0.01% of the total sequence counts. Finally, samples with low counts (< 0.01% of total sequence counts) caused by possible PCR batch failure were also ignored. In total, extraction blanks and NTCs accounted for 332,446 reads (2% of the total data set), of which 99% originated from blanks of one PCR batch. Contamination was likely caused by the primers in the PCR reaction and the bioinformatic pipeline. However, we kept the data from this PCR batch, because we identified sequence types in the blanks that differed significantly from sequence types detected in the lake sediment samples, so they could be removed easily. Likewise, the replicates run on the DNA extractions identified a very similar diversity of sequence types. The final sequence data table, including raw and rarefied abundances, taxonomic assignments and DNA sequence information is provided in PANGEA (https://doi.pangaea.de/10.1594/PANGAEA.917561).
Morphological diatom identification
Twenty-one surface sediment samples used for genetic assessment were also processed for morphological diatom identification. Carbonate and organic components were removed from 0.5 g of sediment by heating with HCl (10%) and H2O2 (30%), respectively. Remaining sediment components were mounted on microscope slides with Naphrax. Up to 500 diatom valves, and not < 250 per slide, were counted at 1000× magnification using a ZEISS microscope equipped with differential interference contrast. Taxonomic identification was based mainly on Krammer and Lange-Bertalot 1986–1991) and additional diatom floras described in Pestryakova et al. (2012). Names of genetically assigned sequence types and morphologically identified taxa are abbreviated for easier visualising in the plots. Full diatom names and their abbreviations in this study are listed in ESM Table S3a and b. Raw and rarefied counts of the microscopic analyses are provided in PANGEA (https://doi.pangaea.de/10.1594/PANGAEA.917561).
Statistical analyses
Statistical analyses were carried out in R 3.4.2 (R Core Team 2017). Calculation of Hill numbers was conducted based on the incidence data using the “iNEXT” package (Hsieh et al. 2016). The diatom composition, based on the read counts of replicates, was similar for each sample (ESM Fig. S1), thus counts of replicates in each sample were summed. To correct for diversity bias caused by different sampling intensity, both genetic read counts and morphological counts were rarefied. Complete R scripts of rarefaction are available at https://github.com/StefanKruse/R_Rarefaction. For the genetic dataset the sampling effort was set as the minimum sample size of observed sequence counts (n = 31,809). This number was used to determine the new sample size of each sample. The original individuals were resampled with 100 repeats to reassign the new rarefied numbers of each sequence type. The final rarefied dataset, used for further statistical analysis, was produced by taking the mean values from the 100 repeats. Sequence types that occurred at ≥ 1% in at least three samples were kept for further statistical analyses. In the morphological dataset, the sampling effort was set as the minimum count size (n = 263), which was used to determine the new total number of counts of each sample. Further rarefaction steps were the same as for the genetic dataset. Diatom taxa that occurred at ≥ 2% in at least three samples were kept for further statistical analyses.
The following statistical analyses were run in the “vegan” package (Oksanen et al. 2013). A double square-root transformation was performed on the relative proportions of the final dataset to reduce the impact of over-represented and rare sequences in the multivariate analysis. The top 20 diatom taxa that had the highest loading on the first axis were included in the ordination plot. Similarity of the genetic- and morphological-based ordination was checked through a symmetric Procrustes analysis and the function protest was used to test for non-randomness (significance) between the two configurations. All environmental variables: maximum depth, Secchi depth, alkalinity, conductivity, dissolved organic carbon (DOC), bicarbonate (HCO3−), Ca2+, SO42−, K+, Sr2+, Al3+, Ba2+, and Fe, but not pH, were log-transformed. Detrended correspondence analysis (DCA) was used to examine the gradient lengths of the diatom dataset. The DCA revealed that the gradient lengths were < 3, suggesting linear relationships between the diatom communities and environmental variables in our study (Ter Braak and Smilauer 2002). Thus, we used redundancy analysis (RDA) as a constrained ordination analysis. Samples from glacial lakes were aggregated in the genetic dataset using the mean within-lake count of samples. Following the RDA, multi-collinearity was tested for by computing variance inflation factors (VIF) for the environmental variables. Variables having a VIF > 10 were removed (O’brien 2007). After each removal, another RDA was run, and the VIFs were re-examined until all VIFs were < 10. Stepwise model selection and a Monte Carlo permutation test were used to keep only significant environmental variables. The significance of each individual component was tested by conditional ordination.
Results
Genetic-based diatom community composition, diversity and diatom-environment relationship
The reference database used for taxonomic assignment resulted in 2039 database entries, of which 1148 sequences (56.3%) were assigned to Bacillariophyta. Among those, 70.5% can be unambiguously distinguished to species level and 77.3% to genus level. All other sequence types can only be assigned to higher taxonomic levels within diatoms. The DNA sequencing of 141 PCR products resulted in 23,628,911 raw sequence reads. After processing with OBITools our total sequencing data contained 15,092,046 paired reads and 5147 sequence types, of which 81.9% belong to Bacillariophyta. After stepwise filtering and de-noising (ESM Table S2), the final dataset of 36 samples contained 10,681,274 reads and 163 unique sequence types (ESM Table S3a).
The most dominant diatom family Fragilariaceae accounts for 47.9% of the total counts, followed by Aulacoseiraceae, with 20.9%. Both families also show the highest diversity within the total dataset. The Fragilariaceae are present with 78 unique sequence types (including Staurosira elliptica, Fragilaria construens and Fragilariaceae) and Aulacoseiraceae account for 34 unique sequence types (including Aulacoseira sp., A. distans var. alpigena, A. subarctica and A. valida). A further 22 sequence types are identified to at least genus level, including Sellaphora, Pinnularia, Amphora, Placoneis, Stauroneis and Urosolenia eriensis. The remaining 29 sequence types are assigned to higher taxonomic levels including Achnanthidiaceae, Cymbellaceae, Bacillariophycidae and Bacillariophyta. Although PCR products were pooled equimolarly, the total number of sequence reads per sample varied substantially (mean 296,702 ± 239,729), with ILI01 having the highest read count (988,126) and KP-03-L11 having the lowest read count (31,809). Rarefaction curves (ESM Fig. S2a) indicate that all samples reached a plateau, indicating sufficient sampling effort for all samples. For further statistical analyses, however, we rarefied the data to the minimum number of sample counts (sample KP-03-L11; 31,809 read counts) and created a normalised dataset.
The distribution of diatom sequence types shows variations according to lake type (Fig. 2a). In glacial lakes Aulacoseira subarctica type 3 is the most dominant taxon and has a mean abundance of 13.89%, followed by Staurosira elliptica type 2 (mean abundance: 12.69%). In thermokarst lakes, S. elliptica type 2 is the most dominant taxon (mean abundance: 13.76%), followed by S. elliptica type 3 (mean abundance: 11.73%). Diatom assemblages from the two lake types overlap, with 146 sequence types occurring in both glacial and thermokarst lakes. Fifteen sequence types occur only in thermokarst lakes and two Aulacoseira types occur only in glacial lakes (ESM Fig. S3a). These two Aulacoseira types (types 2 and 22) occupy only minor fractions, 0.020% and 0.015%, of the glacial lake counts, respectively. The most prominent difference between glacial and thermokarst lakes is the high richness of S. elliptica sequence types in Lake Ilirney (ILI) compared to thermokarst lakes. Regarding the total dataset, samples ILI01, ILI03 and ILI10 have the highest alpha diversity. Staurosira elliptica was present, with 49 reads in glacial lakes, of which 41 occur in Lake Ilirney. In total, 56 reads of S. elliptica were identified in thermokarst lakes. The second most diverse taxon in both lake types is Aulacoseiraceae, which had 31 reads in glacial lakes and 32 in thermokarst lakes. Aulacoseiraceae are highly diverse in northern glacial lakes like Ilirney and Rauchuagytgyn, but the richness drops in Nutenvut samples, and varies among the thermokarst lakes.
Relative abundances (%) of the most dominant genetically retrieved (a) and morphologically identified (b) diatom taxa in glacial and thermokarst lakes from north-eastern Siberia. Blue bars indicate samples from glacial lakes; yellow bars indicate samples from thermokarst lakes. Samples are sorted from north to south in each vegetation zone. Richness within the diatom families Aulacoseiraceae and Fragilariaceae are shown as silhouettes. Hill numbers are given on the right [total species richness (q = 0), the exponent of Shannon's entropy index (q = 1), and the inverse of Simpson's concentration index (q = 2)]. Note the relative abundance scales are different
Our study detected variations in diatom distribution and diversity along the sampled vegetation gradient (Fig. 2a). The diatom community compositional change across the vegetation gradient differs between the lake types. Using the genetic data, we detected major compositional differences among the glacial lakes. The most dominant diatom taxon in tundra glacial Lake Rauchuagytgyn is Stauroneis constricta type 2 (65.4%). Although Lakes Ilirney and Nutenvut are both classified as forest-tundra lakes, the diatom compositions are dominated by Staurosira elliptica type 7 (33.4%) and Bacillariophyta type 4 (48.05%), respectively. In contrast, thermokarst lakes in tundra are dominated by S. elliptica type 3 (52.1%), whereas forest-tundra lakes show the highest abundance of the sequence type Aulacoseira distans var. alpigena (67.3%) and forest lakes have the highest abundance of S. elliptica type 2 (52.8%).
Principal component analysis for the total dataset, including all samples, indicates that the first two PC axes together explain 50.3% of the total variance in the diatom dataset (Fig. 3a). The biplot indicates a separation into two groups of glacial lakes. Toward the positive end of PC2 axis, Lakes Rauchuagytgyn and Ilirney form a glacial tundra/forest-tundra group, whereas Lake Nutenvut forms a glacial forest-tundra group along the negative PC2 axis. Fragilariaceae, Bacillariophyta type 9 and Staurosira elliptica type 34 have high loadings in the Ilirney/Rauchuagytgyn cluster, whereas Bacillariophyta type 4 and Fragilaria construens type 2 are most significant in the Nutenvut cluster. Another cluster in the upper left quadrant consists of thermokarst samples. They are mostly forest lakes with some tundra lakes as well: this cluster has a negative loading, given by Aulacoseira sp. type 1. After aggregating the samples from glacial lakes by taking the mean counts of the samples within each glacial lake, the variance explained by the first two PC axes reduces to 36.6% (ESM Fig. S4a). The first two PC axes jointly explain 41.2% of the diatom variance in the thermokarst lakes (ESM Fig. S4b). The glacial lakes show similar patterns when compared with the non-aggregated PCA by splitting Ilirney/Rauchuagytgyn from Nutenvut. Thermokarst lakes along PC1 show a separation of forest lakes and tundra lakes, whereas along PC2, two clusters of forest-tundra lakes are formed.
We included physical and chemical variables in the RDA to test for relationships between diatom community composition of all lake sites and in-lake environmental conditions. The selected environmental variables have variance inflation factor (VIF) values < 10, suggesting a small inter-set correlation. Further stepwise selection and significance tests indicate that SO42−, DOC, HCO−3 , Secchi depth and K+ are the least collinear and most significant explanatory variables (P < 0.003), with SO42− explaining the highest variance (Table 1, ESM Fig. S6a). The first two constrained axes together explain 42.7% of the variance of the diatom-environment relationship. After aggregating read counts of samples within glacial lakes, SO42− and HCO3− are the most significant environmental variables (P < 0.008) (Table 1, ESM Fig. S6b). When analysing only thermokarst lake samples, the ordination indicates that SO42− and maximum depth are the least inter-set correlated and the most significant explanatory variables (P < 0.05) for diatom assemblages (Table 1, ESM Fig. S6c).
Morphology-based diatom community composition, diversity and diatom-environment relationships
The morphological dataset of 21 samples contains 10,598 counts and is comprised of 176 diatom taxa, of which all were identified to genus or lower taxonomic levels. No diatoms were observed in sample KP-02-L06, thus this sample was excluded from further analyses.
The most dominant diatom family, Fragilariaceae, accounts for 25.6% of the total counts, followed by Aulacoseiraceae, which accounts for 15.5%. The most diverse diatom families are Eunotiaceae (21 taxa), Pinnulariaceae (16 taxa) and Fragilariaceae (16 taxa), whereas the other 25 diatom families possess between one and 14 different taxa. Valve counts varied among the samples (mean 529 ± 78). Sample KP-03-L17 contains the highest total valve count of 697, whereas KP-03-L16 contains the lowest count of 263. Rarefaction curves (ESM Fig. S2b) indicate that all samples reached a plateau, indicating sufficient sampling effort for all samples. Rarefied data were used for further statistical analyses.
The distribution of the morphological diatom data highlights differences between glacial and thermokarst lakes (Fig. 2b). In glacial lakes, Lindavia ocellata is the most dominant diatom taxon (mean abundance: 38.2%), followed by Cyclotella tripartita (mean abundance: 15.2%). In thermokarst lakes Staurosira venter is the most dominant taxon (mean abundance: 20.6%), followed by Tabellaria flocculosa (mean abundance: 16.9%). The diatom communities overlap, with 57 taxa occurring in both lake types (ESM Fig. S3b), but 25 diatom taxa occur only in glacial lakes. The most prominent taxa in relative counts in glacial lakes are Pliocaenicus sp. (5.4%) and Aulacoseira ambigua (1.5%). Similar to the genetic dataset, there are two Aulacoseira taxa that occur exclusively in glacial lakes, A. ambigua and A. perglabra. In thermokarst lakes, we detected 94 unique taxa, with Aulacoseira distans and Staurosira lata having the highest counts, of 3.9% and 2.1%, respectively.
As seen in the genetic data, the glacial lakes are characterised by high diversity of Fragilariaceae, with 9 taxa (Fig. 2b). Highest Fragilariaceae diversity was found in Lake Ilirney, which contains 7 taxa. In thermokarst lakes, Eunotiaceae is the most diverse diatom family, represented by 21 taxa, with high diversity in KP-01-L03, KP-01-L05 and KP-03-L18, which each had 7 taxa. In contrast, only two Eunotiaceae taxa were found in the glacial lakes. The highest overall alpha diversity was found in thermokarst lakes KP-04-L21 and KP-03-L15.
Community compositional change of morphologically identified diatoms across the vegetation gradient, differs between the two lake types. In glacial lakes, Lindavia ocellata is the most dominant taxon in tundra sites (55.1%) and forest-tundra sites (mean abundance 34.01%). In thermokarst lakes, Staurosira venter is the most dominant taxon in tundra sites, which have a mean abundance of 21.7%. In forest-tundra sites, Tabellaria flocculosa is the most dominant taxon (mean abundance 23.1%), whereas in forested sites, Staurosira venter is again the most dominant taxon, with a mean abundance of 61.8%.
Principal component analysis of the total morphologically identified diatom dataset, including all samples, explains 43.2% of the total variance for the first two axes (Fig. 3b). Along PC1, glacial lakes are distinct from thermokarst lakes. The highest loading on PC1 (45.7% of the total variance) is obtained by Lindavia ocellata. Aulacoseira alpigena obtains highest loading on PC2 (26.4% of the total variance). For the thermokarst lakes, the first two PC axes jointly explain 45.9% of the diatom variance (ESM Fig. S5). The highest loading on PC1 is obtained by Staurosirella pinnata (39.3.4% of the total variance). Along PC2, Staurosira venter has the highest loading of 40.6%, and forest lakes are mostly separated from forest-tundra and tundra lakes.
The RDA revealed correlations between the morphologically determined diatom community composition in all the lakes and in-lake water conditions (Table 1, ESM Fig. S7a). The selected environmental variables have VIF values < 10, suggesting a small inter-set correlation. Further stepwise selection and significance testing indicates that HCO3− and DOC are the most significant environmental variables (P < 0.001). The first two constrained axes together explain 32.3% of the variance in the species-environment relationship. Samples from glacial lakes (n = 5) are characterised by low DOC. Thermokarst lakes (n = 15) show increasing HCO3− along the vegetation gradient from tundra to forest lakes. The ordination analysis of just the thermokarst lakes indicates that HCO3− and maximum depth are the least inter-set correlated and the most significant explanatory variables (P < 0.014) for the diatom assemblages (Table 1, ESM Fig. S7b). The forest-tundra lakes from field site 3 are typically deep, whereas samples from field sites 2 and 4 had shallow water depths.
Comparison of spatial diatom patterns obtained from the genetic and morphological approaches
The number of genetically identified sequence types (163) and morphologically identified species (176) is very similar, although the taxonomic assignment of identified sequence types and species shows prominent differences. About 70% of sequence types were identified to lower taxonomic levels (genus or species), whereas the remaining sequence types could only be identified to higher taxonomic levels (family, order, phylum). With the morphological approach, 93.6% of all taxa were identified to species level, with the remaining 7.4% assigned to genus level. In general, both approaches found that Fragilariaceae and Aulacoseiraceae have the highest abundance in the total dataset, but both families show a higher diversity with the genetic approach than with the morphological approach. In particular, the highest diversity identified genetically is in glacial Lake Ilirney, and both approaches similarly indicate that small benthic fragilariods are the most diverse diatom taxon. The highest overall diversity in the morphological dataset is found in thermokarst Lake KP-04-L21.
Procrustes analysis of the genetic-based PCA and morphology-based PCA indicates a statistically significant correlation (P = 0.04) between the two ordinations of samples from all sampling locations. The two ordinations reach best fits (small residual value) between thermokarst lakes like KP-04-L21 (0.12), KP-03-L14 (0.13), and glacial Lakes Ilirney (0.17) and Rauchuagytgygn (ESM Fig. S8). When including only thermokarst lakes, the two ordinations are not significantly similar (P = 0.16).
Both approaches found that HCO3− was the most significant variable (P < 0.008) influencing diatom assemblages at all sampling locations. When assessing thermokarst lake samples only, both approaches find maximum depth to be the most significant variable (P < 0.05).
Discussion
Genetic and morphological diatom composition and diversity
This study combined genetic and morphological approaches to reveal diatom assemblages in surface sediments from glacial and thermokarst lakes along a treeline ecotone in northeastern Siberia. Despite the differences in total sequence types/valve counts and taxonomic resolution, both approaches revealed a similar number of detected taxa and similar diatom patterns across the investigated localities. The genetic approach yielded a higher diversity within specific diatom taxa such as Staurosira sp., Fragilaria sp. and Aulacoseira sp. and found overall higher alpha diversity compared to the morphological approach. The morphological approach enabled better taxonomic identification, to resolve the overall diatom community to species level, which provides more reliable ecological interpretations when exploring the response of diatoms to environmental changes (Dulias et al. 2017). Both approaches revealed dominance in relative abundance of Fragilariaceae and Aulacoseiraceae in the analysed lakes. We are, however, aware that the relative abundance of read counts does not necessarily scale to relative abundances of individuals or biomass in the environmental sample (Deagle et al. 2018). Nevertheless, our genetic findings support findings about the dominance of diatom families determined in our samples by morphologic analysis. In particular, the high abundance of Staurosira and Fragilaria corroborates earlier findings from morphology-based studies of the composition of Siberian diatom communities (Biskaborn et al. 2012; Pestryakova et al. 2012), and is supported by other genetic studies (Stoof-Leichsenring et al. 2014, 2015, 2019; Dulias et al. 2017). Other diatom taxa such as Lindavia ocellata and Tabellaria flocculosa, which are dominant in the morphological dataset, occur rarely in the genetic dataset. It is likely that DNA of planktonic diatoms is under-represented in modern sediments compared to that from benthic diatoms, as DNA of benthic diatoms is better preserved than that from planktonic cells (Dulias et al. 2017). We can’t exclude the influence of methodological issues, like PCR bias, on the findings, especially when using larger PCR cycle numbers (Kelly et al. 2019) and primer preference for some diatom taxa, which might lead to lower overall diversity and over-representation of dominant diatom taxa. In general, the two approaches show large differences in the alpha diversity of diatom taxa. In our study, 56 sequence types were assigned to Staurosira elliptica types in the genetic dataset, but only one taxon was identified by morphology. This high intra-specific variability suggests hidden genetic diversity, which is less detectable with the morphological approach, because of nearly indistinguishable morphotypes of minute fragilariods (Paull et al. 2008; Stoof-Leichsenring et al. 2014). The morphological approach demonstrates highest diversity within the family Eunotiaceae, which is not detected by the genetic approach, or might be hidden behind sequence types of higher taxonomical classifications. Limitations in the taxonomic resolution of the genetic marker and the incompleteness of the genetic database restrict the taxonomic assignment of some sequence types, which is the reason why we used a relatively high threshold for sequence similarity (90%) to the reference database. So far, only a few DNA reference sequences of diatoms from polar freshwaters are available in public databases (e.g. GenBank). Such a deficiency sometimes makes genetic diatom identification ambiguous, especially for polar environments (Ki et al. 2009). Establishing a more up-to-date reference database might be achieved by obtaining sequences of morphologically confirmed specimens from the study area. To overcome the limitations of an insufficient reference database, inferring the molecular taxonomic units directly from eDNA, without reference to taxonomy, might be one alternative approach (Apothéloz-Perret-Gentil et al. 2017).
Diatom composition is affected by lake type and lake water variables
The similarity of ordination analyses obtained from genetic and morphological diatom composition shows that both approaches can separate the glacial lakes from the thermokarst lakes, which results from differences in diatom composition between the two lake types. Most prominent in glacial lakes is the dominance of centric diatoms, such as Aulacoseira sequence types and the species Lindavia ocellata. Aulacoseiraceae, which are heavily silicified diatoms with high sinking rates, are commonly found in deeper Arctic lakes with high water turbulence (Rühland et al. 2003; Buczkó et al. 2010). The high within-lake diversity of Aulacoseira spp. has been identified by genetic and morphological approaches (Risberg et al. 1999; Biskaborn et al. 2019b; Stoof-Leichsenring et al. 2020), and is mostly explained by water depth variations. Lindavia ocellata is known to be dominant in old and large lakes, and the appearance of different morphological variations over glacial/interglacial cycles has been attributed to climate changes or evolutionary selection (Edlund et al. 2003; Cvetkoska et al. 2018). Moreover, both approaches reveal two Aulacoseira taxa that are present solely in glacial lakes, but not in thermokarst lakes, supporting the idea that glacial lakes host endemic/specialised taxa because their prolonged existence enabled evolution to occur (Cvetkoska et al. 2018). Moreover, the genetic data indicate high dominance and diversity of Fragilariaceae in glacial lakes, which may in part be a consequence of the fact that a higher number of within-glacial-lake samples was analysed genetically. Even though glacial Lakes Ilirney and Nutenvut are located in the forest-tundra zone, the diatom composition within Lake Ilirney seems to be more similar to that in adjacent glacial Lake Rauchuagytgyn in the tundra zone. Both the Ilirney/Rauchuagytgyn group and the Nutenvut group have the highest loadings of fragilarioids and sequence types only identified to the taxonomic level of Bacillariophyceae or Bacillariophyta. The dominance of specific fragilarioid sequence types in different lakes presumes the development of some lineages formed by habitat-related specialisation to accommodate lake-type-specific preferences. Furthermore, Fragilarioid taxa are pioneer species that appear immediately after initial formation of a lake, even under severe conditions (Biskaborn et al. 2012). However, in the morphological-based PCA, despite the sparse data, a clear distinction between the Ilirney/Rauchuagytgyn group and the Nutenvut group is missing. This implies that some fragilarioid subspecies and other diatoms within glacial lakes cannot be discriminated by the morphological approach in our data. The genetic-based data reveal clearer differences in the diatom composition of glacial lakes, which could be an indication of in situ evolution of endemic diatom subspecies, and hence suggests a very old age for the origin of these lakes. Current estimates for the age of Lake Ilirney range from about 50 to 60 ka, which would have enabled the evolution of distinct or even endemic lineages within the lake.
The analysed thermokarst lakes show dominance of Fragilariaceae using both analytical approaches. Fragilariceae show the highest diversity using the genetic approach, whereas Eunotiaceae are most diverse using morphological data. Generally, thermokarst lakes are shallow and some of them can freeze to the bottom during severe winters if water depth is less than about 1.5 m, which in our dataset would only affect the shallowest lake, KP-02-L07 (1.7 m). In general, harsh conditions promote high abundances of small benthic fragilarioids. Our results support other studies, which reported that small benthic fragilarioids are more competitive than other species with regard to prolonged ice cover (Lotter et al. 2010), cold temperatures (Laing and Smol 2000; Schmidt et al. 2004), and severe, unstable environmental conditions (Pestryakova et al. 2012, 2018), and are suitable for paleoclimate reconstructions (Finkelstein and Gajewski 2008). The diversity of Eunotia might be a consequence of its specific ecological preferences for some environmental conditions in thermokarst lakes (Pestryakova et al. 2018), however they generally favour acidic conditions and are often found in association with mosses, given their epiphytic mode of life (Michelutti et al. 2007). Moreover, their expansion has been found to be indicative of declining ice cover, associated with recent warming in the Arctic (Wilson et al. 2012).
Based on the genetic data, SO42− concentration is the most influential variable for diatom composition in thermokarst lakes, but is also of primary importance when including glacial lakes. In our dataset, SO42− concentrations form a gradient from very low concentrations in forest and forest-tundra thermokarst lakes, to high concentrations in tundra lakes, and highest concentrations in glacial lakes. Sulfate concentrations are generally low in oligotrophic lakes, however concentrations are higher under oxic conditions, whereas under organic-rich and anoxic conditions SO42− is converted to hydrogen sulfide by sulfate-reducing microbes (Holmer and Storkholm 2001). Kuivila et al. (1989) showed that there is competition between methane-producing and sulfate-reducing bacteria for acetate and hydrogen in Arctic lakes, thus methane and SO42− concentrations show an inverse relationship (Northington and Saros 2016). This could explain why lower SO42− concentrations are seen in deep forest thermokarst lakes, which likely have higher methane production in the underlying thawing permafrost than shallow tundra or glacial lakes, and which were not formed through thermokarst processes.
The second most important variable for the entire set of genetic data (glacial aggregated and thermokarst) is HCO3−, and, for the morphological identifications, DOC. The relevance of HCO3− concurs with a regional study on central Yakutia thermokarst lakes that included water bodies from tundra to dense taiga (Pestryakova 2008). Depending on the vegetation around the lakes, higher evapotranspiration rates in the watersheds lead to higher alkalinity, which is mainly driven by HCO3− concentrations, and likely explains differences in the diatom communities between forested and tundra thermokarst lakes (Herzschuh et al. 2013). The importance of DOC in the lakes analysed in our study was also expected. DOC is responsible for dissolved colour in lakes and limits light transmission through the water column (Scully et al. 1995; Schindler et al. 1996). Higher concentrations of DOC in lakes are believed to come from higher input of dissolved organic matter, which is influenced mainly by landscape properties in the catchment (Bouchard et al. 2016). In our data, we identified an increase in DOC from tundra to forest thermokarst lakes, whereas glacial lakes are generally characterised by very low DOC concentrations and do not show variations in DOC related to vegetation differences. In summary, our data are in good agreement with previous genetic studies, which indicated differences in small fragilarioids in relation to drainage basin vegetation (Stoof-Leichsenring et al. 2015), and concluded that both morphologically and genetically identified diatoms are best explained by the DOC gradient in Siberian lakes along the treeline ecotone (Dulias et al. 2017). Constrained ordinations, excluding glacial lakes, indicate that SO42− and HCO3− are major drivers for thermokarst diatom diversity, with maximum lake depth a secondary variable. Because the thermokarst lakes sampled in our study cover a wide range of maximum depths, from 1.7 to 20 m, water depth and related environmental variables such as light penetration, stratification and turbulence support diatoms with a range of ecological preferences (Pestryakova et al. 2018).
Conclusions
We reported the results of research on surface sediment diatom DNA from glacial and thermokarst lakes in the Chukotka region of northeastern Siberia, Russia, an area that extends across the easternmost expanse of the Siberian treeline ecotone. General agreement between diatom community patterns, using unconstrained and constrained ordinations on genetically and morphologically identified diatom taxa indicates that genetic approaches can be used to infer relationships between diatom assemblage shifts and environmental changes, but are unable to make assignments to lower taxonomic levels. Genetic data resolved detailed sub-taxa variations only for Fragilariaceae and Aulacoseiraceae, which enabled the detection of within-lake diatom patterns in glacial lakes. Our DNA method suggests hidden genetic diversity that is not visible in the morphological data. The generally higher genetic diversity in glacial lakes is likely related to their greater age, range of edaphic settings, and larger variety of lake habitats, which potentially gave rise to endemism. Thermokarst lakes provide highly dynamic small-scale environments, which require stress-tolerant diatom communities that are adapted to the cold Arctic conditions.
This study used DNA in lake sediments to explore modern diatom communities in Arctic lakes and assess their relation to lake type, limnological (physical and chemical) variables, and catchment vegetation. We encourage further genetic investigations of lake sediment archives, which can be used to track the response of diatoms in different lake types to environmental change. In particular, genetic approaches will contribute to understanding how lake attributes affect diatom community development, i.e. loss, gain, and diversity of taxa, and how these attributes will impact diatom communities under continued warming of the Arctic.
References
AMAP (2017) Snow, water, ice and permafrost in the arctic (SWIPA). Arctic Monitoring and Assessment Programme (AMAP), Oslo, Norway, pp 1–269
Apothéloz-Perret-Gentil L, Cordonier A, Straub F, Iseli J, Esling P, Pawlowski J (2017) Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring. Mol Ecol Resour 17:1231–1242. https://doi.org/10.1111/1755-0998.12668
Biskaborn BK, Herzschuh U, Bolshiyanov D, Savelieva L, Diekmann B (2012) Environmental variability in northeastern Siberia during the last ~13,300 year inferred from lake diatoms and sediment-geochemical parameters. Palaeogeogr Palaeoclim Palaeoecol 329:22–36. https://doi.org/10.1016/j.palaeo.2012.02.003
Biskaborn B, Herzschuh U, Bolshiyanov D, Savelieva L, Zibulski R, Diekmann B (2013) Late Holocene thermokarst variability inferred from diatoms in a lake sediment record from the Lena Delta, Siberian Arctic. J Paleolimnol 49:155–170. https://doi.org/10.1007/s10933-012-9650-1
Biskaborn BK, Smith SL, Noetzli J, Matthes H, Vieira G, Streletskiy DA, Schoeneich P, Romanovsky VE, Lewkowicz AG, Abramov A, Allard M, Boike J, Cable WL, Christiansen HH, Delaloye R, Diekmann B, Drozdov D, Etzelmüller B, Grosse G, Guglielmin M, Ingeman-Nielsen T, Isaksen K, Ishikawa M, Johansson M, Johannsson H, Joo A, Kaverin D, Kholodov A, Konstantinov P, Kröger T, Lambiel C, Lanckman J-P, Luo D, Malkova G, Meiklejohn I, Moskalenko N, Oliva M, Phillips M, Ramos M, Sannel ABK, Sergeev D, Seybold C, Skryabin P, Vasiliev A, Wu Q, Yoshikawa K, Zheleznyak M, Lantuit H (2019a) Permafrost is warming at a global scale. Nat Commun 10:1–11. https://doi.org/10.1038/s41467-018-08240-4
Biskaborn BK, Nazarova L, Pestryakova LA, Syrykh L, Funck K, Meyer H, Chapligin B, Vyse S, Gorodnichev R, Zakharov E, Wang R, Schwamborn G, Bailey HL, Diekmann B (2019b) Spatial distribution of environmental indicators in surface sediments of Lake Bolshoe Toko, Yakutia, Russia. Biogeosciences 16:4023–4049. https://doi.org/10.5194/bg-16-4023-2019
Boike J, Georgi C, Kirilin G, Muster S, Abramova K, Fedorova I, Chetverova A, Grigoriev M, Bornemann N, Langer M (2015) Thermal processes of thermokarst lakes in the continuous permafrost zone of northern Siberia - observations and modeling (Lena River Delta, Siberia). Biogeosciences 12:5941–5965. https://doi.org/10.5194/bg-12-5941-2015
Bouchard F, MacDonald LA, Turner KW, Thienpont JR, Medeiros AS, Biskaborn BK, Korosi J, Hall RI, Pienitz R, Wolfe BB (2016) Paleolimnology of thermokarst lakes: a window into permafrost landscape evolution1. Arct Sci 3:91–117. https://doi.org/10.1139/as-2016-0022
Boyer F, Mercier C, Bonin A, Bras Y, Taberlet P, Coissac E (2016) obitools: a unix-inspired software package for DNA metabarcoding. Mol Ecol Resour 16:176–182. https://doi.org/10.1111/1755-0998.12428
Buczkó K, Ognjanova-Rumenova N, Magyari E (2010) Taxonomy, morphology and distribution of some Aulacoseira taxa in glacial lakes in the South Carpathian region. Pol Bot J 55:149–163
Cvetkoska A, Pavlov A, Jovanovska E, Tofilovska S, Blanco S, Ector L, Wagner-Cremer F, Levkov Z (2018) Spatial patterns of diatom diversity and community structure in ancient Lake Ohrid. Hydrobiologia 819:197–215. https://doi.org/10.1007/s10750-018-3637-5
De Barba DM, Miquel C, Boyer F, Mercier C, Rioux D, Coissac E, Taberlet P (2014) DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol Ecol Resour 14:306–323. https://doi.org/10.1111/1755-0998.12188
Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJ, Clare EL, Kartzinel TR, Eveson PJ (2018) Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Mol Ecol 28:391–406. https://doi.org/10.1111/mec.14734
Dulias K, Stoof-Leichsenring KR, Pestryakova LA, Herzschuh U (2017) Sedimentary DNA versus morphology in the analysis of diatom-environment relationships. J Paleolimnol 57:51–66. https://doi.org/10.1007/s10933-016-9926-y
Edlund MB, Willlams RM, Soninkhishig N (2003) The planktonic diatom diversity of ancient Lake Hovsgol, Mongolia. Phycologia 42:232–260. https://doi.org/10.2216/i0031-8884-42-3-232.1
Epp LS, Gussarova G, Boessenkool S, Olsen J, Haile J, Schrøder-Nielsen A, Ludikova A, Hassel K, Stenøien HK, Funder S, Willerslev E, Kjær K, Brochmann C (2015) Lake sediment multi-taxon DNA from North Greenland records early post-glacial appearance of vascular plants and accurately tracks environmental changes. Quat Sci Rev 117:152–163. https://doi.org/10.1016/j.quascirev.2015.03.027
Ficetola G, Coissac E, Zundel S, Riaz T, Shehzad W, Bessière J, Taberlet P, Pompanon F (2010) An In silico approach for the evaluation of DNA barcodes. Bmc Genomics 11:434. https://doi.org/10.1186/1471-2164-11-434
Finkelstein S, Gajewski K (2008) Responses of Fragilarioid-dominated diatom assemblages in a small Arctic lake to Holocene climatic changes, Russell Island, Nunavut, Canada. J Paleolimnol 40:1079–1095. https://doi.org/10.1007/s10933-008-9215-5
Genkal S, Yarushina M (2018) Species of the Genus Nupela Vyverman and Compere (Bacillariophyta) in the Water Bodies of the Far North of Western Siberia and Russian Far East. Int J Algae 20:377–386. https://doi.org/10.1615/interjalgae.v20.i4.40
Grosse G, Jones B, Arp C (2013) Thermokarst lakes, drainage, and drained basins. Thermokarst. https://doi.org/10.1016/b978-0-12-374739-6.00216-5
Gualtieri L, Glushkova O, Brigham-Grette J (2000) Evidence for restricted ice extent during the last glacial maximum in the Koryak Mountains of Chukotka, far eastern Russia. Geol Soc Am Bull 112:1106–1118. https://doi.org/10.1130/0016-7606(2000)112%3c1106:efried%3e2.0.co;2
Guardiola M, Uriz M, Taberlet P, Coissac E, Wangensteen O, Turon X (2015) Deep-sea, deep-sequencing: metabarcoding extracellular DNA from sediments of Marine Canyons. PLoS ONE 10:e0139633. https://doi.org/10.1371/journal.pone.0139633
Herzschuh U, Pestryakova LA, Savelieva LA, Heinecke L, Böhmer T, Biskaborn BK, Andreev A, Ramisch A, Shinneman A, Birks JH (2013) Siberian larch forests and the ion content of thaw lakes form a geochemically functional entity. Nat Commun 4:1–8. https://doi.org/10.1038/ncomms3408
Holmer M, Storkholm P (2001) Sulphate reduction and sulphur cycling in lake sediments: a review. Freshw Biol 46:431–451. https://doi.org/10.1046/j.1365-2427.2001.00687.x
Hsieh T, Ma K, Chao A (2016) iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7:1451–1456. https://doi.org/10.1111/2041-210x.12613
Huang J, Zhang X, Zhang Q, Lin Y, Hao M, Luo Y, Zhao Z, Yao Y, Chen X, Wang L, Nie S, Yin Y, Xu Y, Zhang J (2017) Recently amplified arctic warming has contributed to a continual global warming trend. Nat Clim Change 7:875–879. https://doi.org/10.1038/s41558-017-0009-5
Kelly RP, Shelton AO, Gallego R (2019) Understanding PCR processes to draw meaningful conclusions from environmental DNA studies. Sci Rep 9(1):14. https://doi.org/10.1038/s41598-019-48546-x
Kermarrec L, Franc A, Rimet F, Chaumeil P, Humbert J, Bouchez A (2013) Next-generation sequencing to inventory taxonomic diversity in eukaryotic communities: a test for freshwater diatoms. Mol Ecol Resour 13:607–619. https://doi.org/10.1111/1755-0998.12105
Ki J-S, Cho S-Y, Katano T, Jung S, Lee J, Park B, Kang S-H, Han M-S (2009) Comprehensive comparisons of three pennate diatoms, Diatoma tenuae, Fragilaria vaucheriae, and Navicula pelliculosa, isolated from summer Arctic reservoirs (Svalbard 79°N), by fine-scale morphology and nuclear 18S ribosomal DNA. Polar Biol 32:147–159. https://doi.org/10.1007/s00300-008-0514-0
Krammer K, Lange-Bertalot H (1986–1991) Bacillariophyceae. Gustav Fischer Verlag, Stuttgart, Germany
Kuivila K, Murray J, Devol A, Novelli P (1989) Methane production, sulfate reduction and competition for substrates in the sediments of Lake Washington. Geochim Cosmochim Ac 53:409–416. https://doi.org/10.1016/0016-7037(89)90392-x
Laing TE, Smol JP (2000) Factors influencing diatom distributions in circumpolar treeline lakes of northern Russia. J Phycol 36:1035–1048. https://doi.org/10.1046/j.1529-8817.2000.99229.x
Lenz J, Jones BM, Wetterich S, Tjallingii R, Fritz M, Arp CD, Rudaya N, Grosse G (2016) Impacts of shore expansion and catchment characteristics on lacustrine thermokarst records in permafrost lowlands, Alaska Arctic Coastal Plain. Arktos 2:1–15. https://doi.org/10.1007/s41063-016-0025-0
Lotter AF, Pienitz R, Schmidt R (2010) Diatoms as indicators of environmental change in subarctic and alpine. In: Smol JP, Stoermer EF (eds) The diatoms: applications for the environmental and earth sciences. Cambridge University Press, Cambridge, pp 231–238
Michelutti N, Douglas MS, Smol JP (2007) Evaluating diatom community composition in the absence of marked limnological gradients in the high Arctic: a surface sediment calibration set from Cornwallis Island (Nunavut, Canada). Polar Biol 30:1459–1473. https://doi.org/10.1007/s00300-007-0307-x
Northington RM, Saros JE (2016) Factors controlling methane in Arctic Lakes of Southwest Greenland. PLoS ONE 11:e0159642. https://doi.org/10.1371/journal.pone.0159642
O’brien RM (2007) A caution regarding rules of thumb for variance inflation factors. Qual Quant 41:673–690
Oksanen J, Blanchet GF, Kindt R, Legendre P, Minchin PR, O’hara R, Simpson GL, Solymos P, Stevens HM, Wagner H (2013) Package ‘vegan’. Commun Ecol 2:1–295
Paull TM, Hamilton PB, Gajewski K, LeBlanc M (2008) Numerical analysis of small Arctic diatoms (Bacillariophyceae) representing the Staurosira and Staurosirella species complexes. Phycologia 47:213–224. https://doi.org/10.2216/07-17.1
Pestryakova L (2008) Diatomovye kompleksy ozer Yakutii (Diatom Complexes in the Lakes of Yakutiya). Yakutsk State University, Yakutsk
Pestryakova LA, Herzschuh U, Wetterich S, Ulrich M (2012) Present-day variability and Holocene dynamics of permafrost-affected lakes in central Yakutia (Eastern Siberia) inferred from diatom records. Quat Sci Rev 51:56–70. https://doi.org/10.1016/j.quascirev.2012.06.020
Pestryakova LA, Herzschuh U, Gorodnichev R, Wetterich S (2018) The sensitivity of diatom taxa from Yakutian lakes (north-eastern Siberia) to electrical conductivity and other environmental variables. Polar Res 37:1485625. https://doi.org/10.1080/17518369.2018.1485625
R Core Team (2017) R: A language and environment for statistical computing. R Found Stat Comput Vienna, Austria. https://www.R-project.org/, page R Foundation for Statistical Computing
Risberg J, Sandgren P, Teller JT, Last WM (1999) Siliceous microfossils and mineral magnetic characteristics in a sediment core from Lake Manitoba, Canada: a remnant of glacial Lake Agassiz. Can J Earth Sci 36:1299–1314. https://doi.org/10.1139/e99-022
Rühland KM, Smol JP, Pienitz R (2003) Ecology and spatial distributions of surface-sediment diatoms from 77 lakes in the subarctic Canadian treeline region. Can J Botany 81:57–73. https://doi.org/10.1139/b03-005
Schindler DW, Bayley SE, Parker BR, Beaty KG, Cruikshank DR, Fee EJ, Schindler EU, Stainton MP (1996) The effects of climatic warming on the properties of boreal lakes and streams at the Experimental Lakes Area, northwestern Ontario. Limnol Oceanogr 41:1004–1017. https://doi.org/10.4319/lo.1996.41.5.1004
Schleusner P, Biskaborn BK, Kienast F, Wolter J, Subetto D, Diekmann B (2015) Basin evolution and palaeoenvironmental variability of the thermokarst lake El’gene-Kyuele, Arctic Siberia. Boreas 44:216–229. https://doi.org/10.1111/bor.12084
Schmidt R, Kamenik C, Lange-Bertalot H, Rolf K (2004) Fragilaria and Staurosira (Bacillariophyceae) from sediment surfaces of 40 Lakes in the Austrian Alps in relation to environmental variables, and their potential for palaeoclimatology. J Limnol 63:171–189
Scully N, McQueen D, Lean D, Cooper W (1995) Photochemical formation of hydrogen peroxide in lakes: effects of dissolved organic carbon and ultraviolet radiation. Can J Fish Aquat Sci 52:2675–2681. https://doi.org/10.1139/f95-856
Smol JP, Stoermer EF (2010) The diatoms: applications for the environmental and earth sciences, 2nd edn. Cambridge University Press, Cambridge
Stoof-Leichsenring KR, Epp LS, Trauth MH, Tiedemann R (2012) Hidden diversity in diatoms of Kenyan Lake Naivasha: a genetic approach detects temporal variation. Mol Ecol 21:1918–1930. https://doi.org/10.1111/j.1365-294x.2011.05412.x
Stoof-Leichsenring KR, Bernhardt N, Pestryakova LA, Epp LS, Herzschuh U, Tiedemann R (2014) A combined paleolimnological/genetic analysis of diatoms reveals divergent evolutionary lineages of Staurosira and Staurosirella (Bacillariophyta) in Siberian lake sediments along a latitudinal transect. J Paleolimnol 52:77–93. https://doi.org/10.1007/s10933-014-9779-1
Stoof-Leichsenring KR, Herzschuh U, Pestryakova LA, Klemm J, Epp LS, Tiedemann R (2015) Genetic data from algae sedimentary DNA reflect the influence of environment over geography. Sci Rep-uk 5:1–11. https://doi.org/10.1038/srep12924
Stoof-Leichsenring KR, Epp LS, Pestryakova LA, Herzschuh U (2019) Phylogenetic diversity and environment form assembly rules for Arctic diatom genera—A study on recent and ancient sedimentary DNA. J Biogeogr 00:1–14. https://doi.org/10.1111/jbi.13786
Stoof-Leichsenring KR, Dulias K, Biskaborn BK, Pestryakova LA, Herzschuh U (2020) Lake-depth related pattern of genetic and morphological diatom diversity in boreal Lake Bolshoe Toko, Eastern Siberia. PLoS ONE. https://doi.org/10.1371/journal.pone.0230284
Subetto D, Nazarova L, Pestryakova L, Syrykh L, Andronikov A, Biskaborn B, Diekmann B, Kuznetsov D, Sapelko T, Grekov I (2017) Paleolimnological studies in Russian northern Eurasia: a review. Contemp Probl Ecol 10:327–335. https://doi.org/10.1134/s1995425517040102
Svendsen J, Alexanderson H, Astakhov VI, Demidov I, Dowdeswell JA, Funder S, Gataullin V, Henriksen M, Hjort C, Houmark-Nielsen M, Hubberten HW, Ingólfsson Ó, Jakobsson M, Kjær KH, Larsen E, Lokrantz H, Lunkka J, Lyså A, Mangerud J, Matiouchkov A, Murray A, Möller P, Niessen F, Nikolskaya O, Polyak L, Saarnisto M, Siegert C, Siegert MJ, Spielhagen RF, Stein R (2004) Late Quaternary ice sheet history of northern Eurasia. Quaternary Sci Rev 23:1229–1271. https://doi.org/10.1016/j.quascirev.2003.12.008
Ter Braak CJ, Smilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide: software for canonical community ordination (version 4.5)
Wik M, Varner RK, Anthony K, MacIntyre S, Bastviken D (2016) Climate-sensitive northern lakes and ponds are critical components of methane release. Nat Geosci 9:99–105. https://doi.org/10.1038/ngeo2578
Wilson CR, Michelutti N, Cooke CA, Briner JP, Wolfe AP, Smol JP (2012) Arctic lake ontogeny across multiple interglaciations. Quat Sci Rev 31:112–126. https://doi.org/10.1016/j.quascirev.2011.10.018
Zimmermann J, Glöckner G, Jahn R, Enke N, Gemeinholzer B (2015) Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. Mol Ecol Resour 15:526–542. https://doi.org/10.1111/1755-0998.12336
Acknowledgements
Open Access funding provided by Projekt DEAL. The Russian–German expedition was financed by Grant #5.2711.2017/4.6 from the Russian Foundation for Basic Research (RFBR Grant #18-45-140053 r_a), and the Project of the North-Eastern Federal University (Regulation SMK-P-1/2-242-17 ver. 2.0, order No. 494-OD). We thank Sarah Olischläger for her support in the genetic laboratories and Paul Overduin and Antje Eulenburg for providing the water chemistry data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, S., Herzschuh, U., Pestryakova, L.A. et al. Genetic and morphologic determination of diatom community composition in surface sediments from glacial and thermokarst lakes in the Siberian Arctic. J Paleolimnol 64, 225–242 (2020). https://doi.org/10.1007/s10933-020-00133-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10933-020-00133-1