Abstract
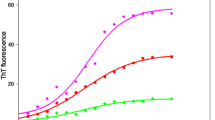
Protein aggregation is related to numerous pathological conditions like Alzheimer’s and Parkinson’s disease. In our study, we have shown that an already existing FDA-approved drug; methotrexate (MTX) can be reprofiled on preformed α-chymotrypsinogen A (α-Cgn A) aggregates. The zymogen showed formation of aggregates upon interaction with mercuric ions, with increasing concentration of Hg2Cl2 (0-150 µM). The hike in ThT and ANS fluorescence concomitant with blue shift, bathochromic shift and the hyperchromic effect in the CR absorbance, RLS and turbidity measurements, substantiate the zymogen β-rich aggregate formation. The secondary structural alterations of α- Cgn A as analyzed by CD measurements, FTIR and Raman spectra showed the transformation of native β-barrel conformation to β-inter-molecular rich aggregates. The native α- Cgn A have about 30% α-helical content which was found to be about 3% in presence of mercuric ions suggesting the formation of aggregates. The amorphous aggregates were visualized by SEM. On incubation of Hg2Cl2 treated α- Cgn A with increasing concentration of the MTX resulted in reversing aggregates to the native-like structure. These results were supported by remarkable decrease in ThT and ANS fluorescence intensities and CR absorbance and also consistent with CD, FTIR, and Raman spectroscopy data. MTX was found to increase the α-helical content of the zymogen from 3 to 15% proposing that drug is efficient in disrupting the β-inter-molecular rich aggregates and reverting it to native like structure. The SEM images are in accordance with CD data showing the disintegration of aggregates. The most effective concentration of the drug was found to be 120 µM. Molecular docking analysis showed that MTX molecule was surrounded by the hydrophobic residues including Phe39, His40, Arg145, Tyr146, Thr151, Gly193, Ser195, and Gly216 and conventional hydrogen bonds, including Gln73 (bond length: 2.67Å), Gly142 (2.59Å), Thr144 (2.81Å), Asn150 (2.73Å), Asp153 (2.71Å), and Cys191 (2.53Å). This investigation will help to find the use of already existing drugs to cure protein misfolding-related abnormalities.






Similar content being viewed by others
Data Availability
Data is provided within the manuscript or supplementary information files.
Abbreviations
- α-Cgn A :
-
α-Chymotrypsinogen A
- Hg2Cl2 :
-
Mercuric chloride
- MTX:
-
Methotrexate
- µM :
-
Micromolar
- ThT:
-
Thioflavin T
- ANS:
-
8-Anilinonaphthalene-1-sulfonic acid
- CR:
-
Congo red
- CD:
-
Circular dichroism
- SEM:
-
Scanning electron microscopy
- FTIR:
-
Fourier transform infrared spectroscopy
- RLS:
-
Rayleigh scattering
References
Ross CA, Poirier MA (2005) What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol 6:891–898. https://doi.org/10.1038/nrm1742
Qadeer A, Zaidi N, Khan RH (2015) Protein folding and aggregation: a revisit of Basic Conception. In: Singh LR, Dar TA, Ahmad P (eds) Proteostasis and Chaperone Surveillance. Springer India, New Delhi, pp 63–87. https://doi.org/10.1007/978-81-322-2467-9_4.
Lee S, Choi MC, Al Adem K, Lukman S, Kim T-Y (2020) Aggregation and Cellular Toxicity of pathogenic or non-pathogenic proteins. Sci Rep 10:5120. https://doi.org/10.1038/s41598-020-62062-3
Taheri R, Hamzkanlu N, Rezvani Y, Niroumand S, Samandar F, Amiri-Tehranizadeh Z, Saberi MR, Chamani J (2022) Exploring the HSA/DNA/lung cancer cells binding behavior of p-Synephrine, a naturally occurring phenyl ethanol amine with anti-adipogenic activity: multi spectroscopic, molecular dynamic and cellular approaches. J Mol Liq 368:120826. https://doi.org/10.1016/j.molliq.2022.120826
Kaffash M, Tolou-Shikhzadeh-Yazdi S, Soleimani S, Hoseinpoor S, Saberi MR, Chamani J (2024) Spectroscopy and molecular simulation on the interaction of Nano-Kaempferol prepared by oil-in-water with two carrier proteins: an investigation of protein–protein interaction. Spectrochim Acta Part A Mol Biomol Spectrosc 309:123815. https://doi.org/10.1016/j.saa.2023.123815
Tabasi M, Maghami P, Amiri-Tehranizadeh Z, Reza Saberi M, Chamani J (2023) New perspective of the ternary complex of nano-curcumin with β-lactoglobulin in the presence of α-lactalbumin: Spectroscopic and molecular dynamic investigations. J Mol Liq 392:123472. https://doi.org/10.1016/j.molliq.2023.123472
Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M (2019) Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18:41–58. https://doi.org/10.1038/nrd.2018.168
Tamás M, Sharma S, Ibstedt S, Jacobson T, Christen P (2014) Heavy Metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 4:252–267. https://doi.org/10.3390/biom4010252
Berlin M (2020) Mercury in dental amalgam: a risk analysis. Neurotoxicology 81:382–386. https://doi.org/10.1016/j.neuro.2020.09.034
Berlin M, Zalups RK, Fowler BA (2015) Mercury. Handbook on the Toxicology of metals. Elsevier, pp 1013–1075. https://doi.org/10.1016/B978-0-444-59453-2.00046-9.
Wang D, Bode W, Huber R (1985) Bovine Chymotrypsinogen J Mol Biology 185:595–624. https://doi.org/10.1016/0022-2836(85)90074-9
DuBay KF, Pawar AP, Chiti F, Zurdo J, Dobson CM, Vendruscolo M (2004) Prediction of the Absolute Aggregation Rates of Amyloidogenic Polypeptide Chains. J Mol Biol 341:1317–1326. https://doi.org/10.1016/j.jmb.2004.06.043
Fairbanks LD, RüCKEMANN K, Qiu Y, Hawrylowicz CM, Richards DF, Swaminathan R, Kirschbaum B, Simmonds HA (1999) Methotrexate inhibits the first committed step of purine biosynthesis in mitogen-stimulated human T-lymphocytes: a metabolic basis for efficacy in rheumatoid arthritis? Biochem J 342:143–152. https://doi.org/10.1042/bj3420143
Puig L (2014) Metotrexato: novedades terapéuticas. Actas Dermosifiliogr 105:583–589. https://doi.org/10.1016/j.ad.2012.11.017
Gray O, McDonnell GV, Forbes RB (2010) Methotrexate for multiple sclerosis. Cochrane Database Syst Reviews 2004. https://doi.org/10.1002/14651858.CD003208.pub2
Malek-Esfandiari Z, Rezvani-Noghani A, Sohrabi T, Mokaberi P, Amiri-Tehranizadeh Z, Chamani J (2023) Molecular Dynamics and Multi-spectroscopic of the Interaction behavior between bladder Cancer cells and calf Thymus DNA with Rebeccamycin: apoptosis through the Down Regulation of PI3K/AKT signaling pathway. J Fluoresc 33:1537–1557. https://doi.org/10.1007/s10895-023-03169-4
Lintner K, Fermandjian S, St. Pierre S, Regoli D (1979) Proton NMR study of the conformation of bradykinin: pH titration. Biochem Biophys Res Commun 91:803–811. https://doi.org/10.1016/0006-291X(79)91951-X
Abidi M, Iram A, Furkan M, Naeem A (2017) Secondary structural alterations in glucoamylase as an influence of protein aggregation. Int J Biol Macromol 98:459–468. https://doi.org/10.1016/j.ijbiomac.2017.01.086
Stryer L (1968) Fluorescence spectroscopy of proteins: fluorescent probes provide insight into the structure, interactions, and dynamics of proteins. Science 162:526–533. https://doi.org/10.1126/science.162.3853.526
Vernaglia BA, Huang J, Clark ED (2004) Guanidine hydrochloride can induce amyloid fibril formation from Hen Egg-White Lysozyme. Biomacromolecules 5:1362–1370. https://doi.org/10.1021/bm0498979
Moosavi-Movahedi AA, Chamani J, Ghourchian H, Shafiey H, Sorenson CM, Sheibani N (2003) Electrochemical evidence for the molten Globule States of Cytochrome c Induced by N-Alkyl sulfates at low concentrations. J Protein Chem 22:23–30. https://doi.org/10.1023/A:1023011609931
Simon LM, Kotormán M, Garab G, Laczkó I (2001) Structure and activity of α-Chymotrypsin and trypsin in Aqueous Organic Media. Biochem Biophys Res Commun 280:1367–1371. https://doi.org/10.1006/bbrc.2001.4282
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935–949. https://doi.org/10.1038/nrd1549
Lu C-H, Chen C-C, Yu C-S, Liu Y-Y, Liu J-J, Wei S-T, Lin Y-F (2022) MIB2: metal ion-binding site prediction and modeling server. Bioinformatics 38:4428–4429. https://doi.org/10.1093/bioinformatics/btac534
Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416:507–511. https://doi.org/10.1038/416507a
Stefani M, Dobson CM (2003) Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med 81:678–699. https://doi.org/10.1007/s00109-003-0464-5
Mathew M, Aravindakumar DTVCT, Aravind UK (2021) Potential involvement of environmental triggers in protein aggregation with mercuric chloride as a model. Int J Biol Macromol 174:153–161. https://doi.org/10.1016/j.ijbiomac.2021.01.134
Meleleo D, Sblano C, Storelli MM, Mallamaci R (2020) Evidence of cadmium and mercury involvement in the Aβ42 aggregation process. Biophys Chem 266:106453. https://doi.org/10.1016/j.bpc.2020.106453
Fazili NA, Naeem A (2015) Anti-fibrillation potency of caffeic acid against an antidepressant induced fibrillogenesis of human α-synuclein: implications for Parkinson’s disease. Biochimie 108:178–185. https://doi.org/10.1016/j.biochi.2014.11.011
Perumal S, Dubey K, Badhwar R, George KJ, Sharma RK, Bagler G, Madhan B, Kar K (2015) Capsaicin inhibits collagen fibril formation and increases the stability of collagen fibers. Eur Biophys J 44:69–76. https://doi.org/10.1007/s00249-014-1002-9
Haran G (2012) How, when and why proteins collapse: the relation to folding. Curr Opin Struct Biol 22:14–20. https://doi.org/10.1016/j.sbi.2011.10.005
Voropai ES, Samtsov MP, Kaplevskii KN, Maskevich AA, Stepuro VI, Povarova OI, Kuznetsova IM, Turoverov KK, Fink AL, Uverskii VN (2003) Spectral properties of Thioflavin T and its complexes with amyloid fibrils. J Appl Spectrosc 70:868–874. https://doi.org/10.1023/B:JAPS.0000016303.37573.7e
Gasymov OK, Glasgow BJ (2007) ANS fluorescence: potential to augment the identification of the external binding sites of proteins, Biochimica et Biophysica Acta (BBA) - proteins and proteomics 1774 403–411. https://doi.org/10.1016/j.bbapap.2007.01.002
Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas’ AF, Gilmanshin RI (1991) Study of the ?Molten globule? Intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 31:119–128. https://doi.org/10.1002/bip.360310111
Hawe A, Sutter M, Jiskoot W (2008) Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res 25:1487–1499. https://doi.org/10.1007/s11095-007-9516-9
Lindgren M, Sörgjerd K, Hammarström P (2005) Detection and characterization of aggregates, Prefibrillar Amyloidogenic Oligomers, and Protofibrils using fluorescence spectroscopy. Biophys J 88:4200–4212. https://doi.org/10.1529/biophysj.104.049700
Vetri V, Canale C, Relini A, Librizzi F, Militello V, Gliozzi A, Leone M (2007) Amyloid fibrils formation and amorphous aggregation in concanavalin A. Biophys Chem 125:184–190. https://doi.org/10.1016/j.bpc.2006.07.012
Groenning M (2010) Binding mode of thioflavin T and other molecular probes in the context of amyloid fibrils—current status. J Chem Biol 3:1–18. https://doi.org/10.1007/s12154-009-0027-5
Wu C, Scott J, Shea J-E (2012) Binding of Congo Red to amyloid protofibrils of the Alzheimer Aβ9–40 peptide probed by Molecular Dynamics simulations. Biophys J 103:550–557. https://doi.org/10.1016/j.bpj.2012.07.008
Khatua DK, Halder M (2019) Distinctively complete inhibition of fibrillation of serum albumins by methotrexate in vitro: experimental and modelling studies to understand the tuning of protein misfolding-related aggregations. New J Chem 43:18983–18987. https://doi.org/10.1039/C9NJ05128G
Chamani J, Heshmati M (2008) Mechanism for stabilization of the molten globule state of papain by sodium n-alkyl sulfates: Spectroscopic and calorimetric approaches. J Colloid Interface Sci 322:119–127. https://doi.org/10.1016/j.jcis.2008.03.001
Ramshini H, Parrini C, Relini A, Zampagni M, Mannini B, Pesce A, Saboury AA, Nemat-Gorgani M, Chiti F (2011) Large proteins have a great tendency to Aggregate but a low propensity to form amyloid fibrils. PLoS ONE 6:e16075. https://doi.org/10.1371/journal.pone.0016075
Matsuo K (2004) Secondary-structure analysis of proteins by Vacuum-Ultraviolet Circular Dichroism Spectroscopy. J BioChem 135:405–411. https://doi.org/10.1093/jb/mvh048
De Diego T, Lozano P, Gmouh S, Vaultier M, Iborra JL (2004) Fluorescence and CD spectroscopic analysis of the ?-chymotrypsin stabilization by the ionic liquid, 1-ethyl-3-methylimidazolium bis[(trifluoromethyl)sulfonyl]amide, Biotechnol. Bioeng 88:916–924. https://doi.org/10.1002/bit.20330
Yamaguchi K, Matsumoto T, Kuwata K (2008) Critical region for amyloid fibril formation of mouse prion protein: unusual Amyloidogenic properties of the Helix 2 peptide. Biochemistry 47:13242–13251. https://doi.org/10.1021/bi801562w
Kuhar N, Sil S, Umapathy S (2021) Potential of Raman spectroscopic techniques to study proteins. Spectrochim Acta Part A Mol Biomol Spectrosc 258:119712. https://doi.org/10.1016/j.saa.2021.119712
Herrero AM (2008) Raman Spectroscopy for Monitoring Protein Structure in Muscle Food Systems. Crit Rev Food Sci Nutr 48:512–523. https://doi.org/10.1080/10408390701537385
Friedman R (2022) Computational studies of protein–drug binding affinity changes upon mutations in the drug target. WIREs Comput Mol Sci 12:e1563. https://doi.org/10.1002/wcms.1563
Moelbert S, Emberly E, Tang C (2004) Correlation between sequence hydrophobicity and surface-exposure pattern of database proteins. Protein Sci 13:752–762. https://doi.org/10.1110/ps.03431704
Millucci L, Spreafico A, Tinti L, Braconi D, Ghezzi L, Paccagnini E, Bernardini G, Amato L, Laschi M, Selvi E, Galeazzi M, Mannoni A, Benucci M, Lupetti P, Chellini F, Orlandini M, Santucci A (2012) Alkaptonuria is a novel human secondary amyloidogenic disease. Biochim et Biophys Acta (BBA) - Mol Basis Disease 1822:1682–1691. https://doi.org/10.1016/j.bbadis.2012.07.011
Hamed KM, Dighriri IM, Baomar AF, Alharthy BT, Alenazi FE, Alali GH, Alenazy RH, Alhumaidi NT, Alhulayfi DH, Alotaibi YB, Alhumaidan SS, Alhaddad ZA, Humadi AA, Alzahrani SA, Alobaid RH (2022) Overview of Methotrexate Toxicity: a Comprehensive Literature Review. Cureus. https://doi.org/10.7759/cureus.29518
Abolmaali SS, Tamaddon AM, Dinarvand R (2013) A review of therapeutic challenges and achievements of methotrexate delivery systems for treatment of cancer and rheumatoid arthritis. Cancer Chemother Pharmacol 71:1115–1130. https://doi.org/10.1007/s00280-012-2062-0
Batchelor T, Carson K, O’Neill A, Grossman SA, Alavi J, New P, Hochberg F, Priet R (2003) Treatment of primary CNS lymphoma with methotrexate and deferred Radiotherapy: a report of NABTT 96–07, JCO 21 1044–1049. https://doi.org/10.1200/JCO.2003.03.036
Acknowledgements
The authors are highly thankful for the facilities provided by the Department of Biochemistry, Faculty of Life Sciences, AMU, Aligarh. The authors acknowledged the Department of Chemistry, AMU, Aligarh for providing FTIR spectroscopy and USIF, AMU for providing SEM and Raman spectroscopy facility. N. K. A. is a recipient of MANF- JRF funded by the Ministry of Minority Affairs, India.
Funding
No external funding was received for conducting this work.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: N. K. A. and A. N. Performed the experiments: N. K. A. and A. R. Analysed the data: N.K. A. and A. N. Wrote the paper: N. K. A., A.R., and A. N. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ansari, N.K., Rais, A. & Naeem, A. Methotrexate for Drug Repurposing as an Anti-Aggregatory Agent to Mercuric Treated α-Chymotrypsinogen-A. Protein J (2024). https://doi.org/10.1007/s10930-024-10187-z
Accepted:
Published:
DOI: https://doi.org/10.1007/s10930-024-10187-z