Abstract
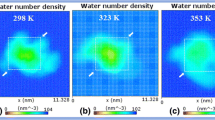
The present study aims at understanding the effect of organic solvents on the specific proteolytic activity and operational stability of asclepain cI in aqueous-organic media, using correlations between geometrical and structural parameters of asclepain cI. These correlations were determined by molecular dynamics (MD) simulations and the secondary structure of the enzyme validated by Fourier-transform Infrared (FTIR) spectroscopy. Asclepain cI exhibited significantly higher catalytic potential in 29 of the 42 aqueous-organic media tested, composed by 0.1 mM TRIS hydrochloride buffer pH 8 (TCB) and an organic solvent, than in buffer alone. Asclepain cI in water-organic miscible systems showed high FTIR spectral similarity with that obtained in TCB, while in immiscible systems the enzyme acquired different secondary structures than in buffer. Among the conditions studied, asclepain cI showed the highest catalytic potential in 50% v/v ethyl acetate in TCB. According to MD simulations, that medium elicited solvation and flexibility changes around the active center of asclepain cI and conducted to a new secondary structure with the active center preserved. These results provide valuable insights into the elucidation of the molecular mechanism of asclepain cI tolerance to organic solvents and pave the way for its future application for the synthesis of peptides in aqueous-organic media.
Graphical Abstract











Similar content being viewed by others
References
Obregón WD, Liggieri CS, Trejo SA, Avilés FX, Vairo-Cavalli SE, Priolo NS (2009) Characterization of papain-like isoenzymes from latex of Asclepias curassavica by molecular biology validated by proteomic approach. Biochimie. https://doi.org/10.1016/j.biochi.2009.07.017
Gurumallesh P, Alagu K, Ramakrishnan B, Muthusamy S (2019) A systematic reconsideration on proteases. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2019.01.081
Origone A, Barberis S, Illanes A, Guzmán F, Camí G, Liggieri C, Martínez R, Bernal C (2020) Improvement of enzymatic performance of Asclepias curassavica L. proteases by immobilization. Application to the synthesis of an antihypertensive peptide. Process Biochem. https://doi.org/10.1016/j.procbio
Kumar A, Dhar K, Kanwar SS, Arora PK (2016) Lipase catalysis in organic solvents: advantages and applications. Biol Proced Online. https://doi.org/10.1186/s12575-016-0033-2
Devi NA, Radhika GB, Bhargavi RJ (2017) Lipase catalyzed transesterification of ethyl butyrate synthesis in n-hexane—a kinetic study. Technol J Food Sci. https://doi.org/10.1007/s13197-017-2725-2
Taher H, Al-Zuhair S (2017) The use of alternative solvents in enzymatic biodiesel production: a review. Biofuel Bioprod Bior. https://doi.org/10.1002/bbb.1727
Guzmán F, Barberis S, Illanes A (2007) Peptide synthesis: chemical or enzymatic. Review article. J Biotechnol Electron. https://doi.org/10.2225/vol10-issue2-fulltext-13
Grundtvig IPR, Heintz S, Krühne U, Gernaey KV, Adlercreutz P, Hayler JD, Wells AS, Woodley JM (2018) Screening of organic solvents for bioprocesses using aqueous-organic two-phase systems. Review. Biotechnol Adv. https://doi.org/10.1016/j.biotechadv.2018.05.007
Priyanka P, Tan Y, Kinsella GK, Henehan GT, Ryan BJ (2019) Solvent stable microbial lipases: current understanding and biotechnological applications. Biotechnol Lett. https://doi.org/10.1007/s10529-018-02633-7
Yang L, Dordick JS, Garde S (2004) Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophys J. https://doi.org/10.1529/biophysj.104.041269
Quiroga E, Priolo N, Marchese J, Barberi S (2006) Behaviour of araujiain, a new cysteine phytoprotease, in organic media with low water content. Electron J Biotechnol. https://doi.org/10.2225/vol9-issue1-fulltext-6
Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz Jr KM, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem. https://doi.org/10.1002/jcc.20290
Baltacioglu H, Bayindirli A, Severcan M, Severcan F (2015) Effect of thermal treatment on secondary structure and conformational change of mushroom polyphenol oxidase (PPO) as food quality related enzyme: a FTIR study. Food Chem. https://doi.org/10.1016/j.foodchem.2015.04.097
Usoltsev D, Sitnikova VE, Kajava A, Uspenskaya M (2019) Systematic FTIR spectroscopy study of the secondary structure changes in human serum albumin under various denaturation conditions. Biomolecules. https://doi.org/10.3390/biom9080359
Yu SW, Rao SS (2014) Advances in the management of constipation-predominant irritable bowel syndrome: the role of linaclotide. Therap Adv Gastroenterol. https://doi.org/10.1177/1756283X14537882
Fosgerau K, Hoffmann T (2015) Peptide therapeutics: current status and future directions. Drug Discov Today. https://doi.org/10.1016/j.drudis.2014.10.003
Zhang L, Hapon MB, Goyeneche AA, Srinivasan R, Gamarra-Luques CD, Callegari EA, Drappeau DD, Terpstra EJ, Pan B, Knapp JR, Chien J, Wang X, Eyster KM, Telleria CM (2016) Mifepristone increases mRNA translation rate, triggers the unfolded protein response, increases autophagic flux, and kills ovarian cancer cells in combination with proteasome or lysosome inhibitors. Mol Oncol. https://doi.org/10.1016/j.molonc.2016.05.001
Chinnadurai RK, Khan N, Meghwanshi GK, Ponne S, Althobiti M, Rajender Kumar R (2023) Review. Current research status of anti-cancer peptides: mechanism of action, production, and clinical applications. Biomed Pharmacother. https://doi.org/10.1016/j.biopha.2023.114996
López-García G, Dublan-García O, Arizmendi-Cotero D, Gómez Oliván LM (2022) Antioxidant and antimicrobial peptides derived from food proteins. Molecules. https://doi.org/10.3390/molecules27041343
Mahlapuu M, Björn C, Jonas Ekblom J (2020) Review Articles. Antimicrobial peptides as therapeutic agents: opportunities and challenges. Crit Rev Biotechnol. https://doi.org/10.1080/07388551.2020.1796576
Reyes Jara AM, Liggieri CS, Bruno MA (2018) Preparation of soy protein hydrolysates with antioxidant activity by using peptidases from latex of Maclura pomifera fruits. Food Chem. https://doi.org/10.1016/j.foodchem.2018.05.013
Liggieri C, Obregón W, Trejo S, Priolo N (2009) Biochemical analysis of a papain-like protease isolated from the latex of Asclepias curassavica L. Acta Biochim Biophy Sin. https://doi.org/10.1093/abbs/gmn018
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal Biochem. https://doi.org/10.1016/0003-2697(76)90527-3
Abraham MH, Grellier PL, Abboud JLM, Doherty RH, Taft RW (1988) Solvent effects in organic chemistry-recent developments. Can J Chem. https://doi.org/10.1139/v88-420
Abraham MH, Lieb WR, Franks NP (1991) Role of hydrogen bonding in general anesthesia. J Pharm Sci. https://doi.org/10.1002/jps.2600800802
Abraham MH, McGowan JC (1987) The use of characteristic volumes to measure cavity terms in reversed phase liquid chromatography. Chromatographia. https://doi.org/10.1007/BF02311772
Reichardt C (1994) Solvatochromic dyes as solvent polarity indicators. Chem Rev. https://doi.org/10.1021/cr00032a005
Abboud J-LM, Notari R (1999) Critical compilation of scales of solvent parameters. Part I. Pure, non-hydrogen bond donor solvents. Pure Appl Chem. https://doi.org/10.1351/pac199971040645
Barberis S, Quiroga E, Morcelle S, Priolo N, Luco JM (2006) Study of phytoproteases stability in aqueous-organic biphasic systems using linear free energy relationships. J Mol Catal B. https://doi.org/10.1016/jmolcatb.2005.11.011
Urrutia P, Bernal C, Wilson L, Illanes A (2018) Use of chitosan heterofunctionality for enzyme immobilization: β-galactosidase immobilization for galacto-oligosaccharide synthesis. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2018.04.112
Meyer JD, Manning MC, Carpenter JF (2004) Effects of potassium bromide disk formation on the infrared spectra of dried model proteins. J Pharm Sci. https://doi.org/10.1002/jps.10562
Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins. https://doi.org/10.1002/prot.340230412
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. https://doi.org/10.1002/bip.360221211
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. https://doi.org/10.1038/nprot.2015.053
Martínez L, Andrade R, Birgin EG, Martínez JM (2009) Packmol: a package for building initial configurations for molecular dynamics simulations. J Comp Chem. https://doi.org/10.1002/jcc.21224
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. https://doi.org/10.1002/prot.21123
Andrews J, Blaisten-Barojas E (2019) Exploring with molecular dynamics the structural fate of PLGA oligomers in various solvents. J Phys Chem B. https://doi.org/10.1021/acs.jpcb.9b06681
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys. https://doi.org/10.1021/acs.jpcb.9b06681
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys. https://doi.org/10.1063/1.470117
Hess B (2008) P-LINCS: a parallel linear constraint solver for molecular simulation. J Chem Theory Comput. https://doi.org/10.1021/ct700200b
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys doi 10(1063/1):2408420
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. https://doi.org/10.1063/1.328693
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. https://doi.org/10.1016/j.softx.2015.06.001
Martínez L, Andreani R, Martínez JM (2007) Convergent algorithms for protein structural alignment. BMC Bioinform. https://doi.org/10.1186/1471-2105-8-306
Andreani R, Martínez JM, Martínez L, Yano F (2008) Continuous optimization methods for structural alignment. Math Program. https://doi.org/10.1007/s10107-006-0091-3
Martínez L (2015) Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE. https://doi.org/10.1371/journal.pone.0119264
Barth A (2007) Infrared spectroscopy of proteins. Biochim Biophys Acta. https://doi.org/10.1016/j.bbabio.2007.06.004
Murayama K, Tomida M (2004) Heat-induced secondary structure and conformation change of bovine serum albumin investigated by Fourier transform infrared spectroscopy. Biochemistry. https://doi.org/10.1021/bi0489154
Marcelino AMC, Gierasch LM (2008) Roles of β-turns in protein folding: from peptide models to protein engineering. Biopolymers. https://doi.org/10.1002/bip.20960
Lobanov MIu, Bogatyreva NS, Galzitskaia OV (2008) Radius of gyration is indicator of compactness of protein structure. Mol Biol (Mosk) 42(4):701–706 (in Russian)
Prestrelski SJ, Tedeschi N, Arakawa T, Carpenter JF (1993) Dehydration induced conformational transitions in proteins and their inhibition by stabilizers. Biophys J. https://doi.org/10.1016/S0006-3495(93)81120-2
Kendrick BS, Dong A, Allison SD, Manning MC, Carpenter JF (1996) Quantitation of the area of overlap between second-derivative amide I infrared spectra to determine the structural similarity of a protein in different states. J Pharm Sci. https://doi.org/10.1021/js950332f
Shivu B, Seshadri S, Li J, Oberg KA, Uversky VN, Fink AL (2013) Distinct β-sheet structure in protein aggregates determined by ATR-FTIR spectroscopy. Biochemistry. https://doi.org/10.1021/bi400625v
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature. https://doi.org/10.1038/s41586-021-03819-2
Varadi M, Anyango S, Deshpande M et al (2022) AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab1061
Schrödinger LLC (2015) The PyMOL molecular graphics system, version 1.8
Abusham RAK, Masomian M, Salleh AB, Leow ATC, Rahman RNZRA (2019) An in-silico approach to understanding the structure-function: a molecular dynamics simulation study of rand serine protease properties form Bacillus subtilis in aqueous solvents. Adv Biotechnol Microbiol. https://doi.org/10.19080/AIBM.2019.12.555828
Gu Z, Lai JL, Hang J, Zhang C, Wang S, Jiao Y, Liu S, Fang Y (2019) Theoretical and experimental studies on the conformational changes of organic solvent-stable protease from Bacillus sphaericus DS11 in methanol/water mixtures. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2019.01.196
Acknowledgements
The authors thank Dr. Sandra Signorella at IQUIR-CONICET-Rosario by the FTIR facilities and Dr. Estela Blaisten-Barojas that kindly provided the ethyl ester of acetic acid force field parameters. Anabella Lucía Origone is postdoctoral scholar at the National Council of Scientific and Technique Research (CONICET), Argentina. Esteban Vega Hissi and Sonia Barberis are scientific researchers at CCT-San Luis-CONICET, San Luis, Argentina.
Funding
This work was supported by the National University of San Luis, San Luis, Argentina (Grant Number 2-0718, 2018-2022).
Author information
Authors and Affiliations
Contributions
SB designed the experiments, did the data analysis together with AO, and wrote the manuscript. AO did the experimental trials, data collection and analysis. EVH performed molecular dynamics simulations. GC did the FTIR spectra. CL prepared the purified enzyme extracts. AI collaborated with SB in reviewing the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare they have no financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Origone, A.L., Hissi, E.G.V., Liggieri, C.S. et al. Effect of Organic Solvents on the Activity, Stability and Secondary Structure of asclepain cI, Using FTIR and Molecular Dynamics Simulations. Protein J (2024). https://doi.org/10.1007/s10930-024-10182-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s10930-024-10182-4