Abstract
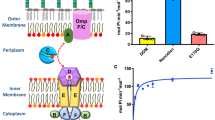
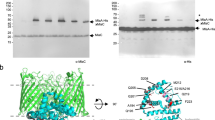
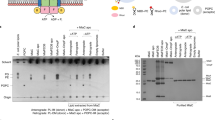
The membrane-associated solute-binding protein (SBP) MlaD of the maintenance of lipid asymmetry (Mla) system has been reported to help the transport of phospholipids (PLs) between the outer and inner membranes of Gram-negative bacteria. Despite the availability of structural information, the molecular mechanism underlying the transport of PLs and the ancestry of the protein MlaD remain unclear. In this study, we report the crystal structures of the periplasmic region of MlaD from Escherichia coli (EcMlaD) at a resolution range of 2.3–3.2 Å. The EcMlaD protomer consists of two distinct regions, viz. N-terminal β-barrel fold consisting of seven strands (referred to as MlaD domain) and C-terminal α-helical domain (HD). The protein EcMlaD oligomerizes to give rise to a homo-hexameric ring with a central channel that is hydrophobic and continuous with a variable diameter. Interestingly, the structural analysis revealed that the HD, instead of the MlaD domain, plays a critical role in determining the oligomeric state of the protein. Based on the analysis of available structural information, we propose a working mechanism of PL transport, viz. “asymmetric protomer movement (APM)”. Wherein half of the EcMlaD hexamer would rise in the periplasmic side along with an outward movement of pore loops, resulting in the change of the central channel geometry. Furthermore, this study highlights that, unlike typical SBPs, EcMlaD possesses a fold similar to EF/AMT-type beta(6)-barrel and a unique ancestry. Altogether, the findings firmly establish EcMlaD to be a non-canonical SBP with a unique ligand-transport mechanism.





Similar content being viewed by others
Data Availability
All the data are submitted along with the manuscript. The three-dimensional atomic coordinates and the structure factors have been deposited in the RCSB Protein Data Bank with the accession codes 8HQA, 8HPZ and 8HQ9.
Abbreviations
- ABC:
-
ATP-binding cassette
- ATP:
-
adenosine triphosphate
- APM:
-
asymmetric protomer movement
- CTD:
-
C-terminal domain
- IF:
-
interfacial
- IM:
-
inner membrane
- LPS:
-
lipopolysaccharide
- LTP:
-
lipid-transfer proteins
- Mla:
-
maintenance of lipid asymmetry
- NTD:
-
N-terminal domain
- OM:
-
outer membrane
- PDB:
-
protein data bank
- PEF:
-
phosphatidylethanolamine
- PL:
-
phospholipid
- PLP:
-
pore loop
- RMSD:
-
root-mean-square deviation
- SBP:
-
solute-binding protein
- SDM:
-
segmented domain movement
- TMD:
-
transmembrane domain
References
Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656
Delcour AH (2009) Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta - Proteins Proteom 1794:808–816
Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, Herrera CM, Trent MS (2016) The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol 70:255–278
Bishop RE (2008) Structural biology of membrane-intrinsic β-barrel enzymes: sentinels of the bacterial outer membrane. Biochim Biophys Acta - Biomembr 1778:1881–1896
Malinverni JC, Silhavy TJ (2009) An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci USA 106:8009–8014
Hughes GW, Hall SC, Laxton CS, Sridhar P, Mahadi AH, Hatton C, Piggot TJ, Wotherspoon PJ, Leney AC, Ward DG, Jamshad M, Spana V, Cadby IT, Harding C, Isom GL, Bryant JA, Parr RJ, Yakub Y, Jeeves M, Huber D, Henderson IR, Clifton LA, Lovering AL, Knowles TJ (2019) Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat Microbiol 4:1692–1705
Coudray N, Isom GL, MacRae MR, Saiduddin MN, Bhabha G, Ekiert DC (2020) Structure of bacterial phospholipid transporter MlaFEDB with substrate bound. eLife 9:e62518
Low WY, Thong S, Chng SS (2021) ATP disrupts lipid-binding equilibrium to drive retrograde transport critical for bacterial outer membrane asymmetry. Proc Natl Acad Sci USA 118:e2110055118
Ekiert DC, Coudray N, Bhabha G (2022) Structure and mechanism of the bacterial lipid ABC transporter, MlaFEDB. Curr Opin Struct Biol 76:102429
Rees DC, Johnson E, Lewinson O (2009) ABC transporters: the power to change. Nat Rev Mol Cell Biol 10:218–227
Wilkens S (2015) Structure and mechanism of ABC transporters. F1000Prime Rep 7:14
Maqbool A, Horler RS, Muller A, Wilkinson AJ, Wilson KS, Thomas GH (2015) The substrate-binding protein in bacterial ABC transporters: dissecting roles in the evolution of substrate specificity. Biochem Soc Trans 43:1011–1017
Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B (2010) A structural classification of substrate-binding proteins. FEBS Lett 584:2606–2617
Fukami-Kobayashi K, Tateno Y, Nishikawa K (1999) Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J Mol Biol 286:279–290
Lee YH, Deka RK, Norgard MV, Radolf JD, Hasemann CA (1999) Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat Struct Mol Biol 6:628–633
Scheepers GH, Lycklama a Nijeholt JA, Poolman B (2016) An updated structural classification of substrate-binding proteins. FEBS Lett 590:4393–4401
Chandravanshi M, Tripathi SK, Kanaujia SP (2021) An updated classification and mechanistic insights into ligand binding of the substrate-binding proteins. FEBS Lett 595:2395–2409
Dutta A, Kanaujia SP (2022) MlaC belongs to a unique class of non-canonical substrate-binding proteins and follows a novel phospholipid-binding mechanism. J Struct Biol 214:107896
Wong LH, Gatta AT, Levine TP (2019) Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat Rev Mol Cell Biol 20:85–101
Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW (1993) Cloning of an M. Tuberculosis DNA fragment associated with entry and survival inside cells. Science 261:1454–1457
Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD (2017) Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285
Dutta A, Chandravanshi M, Kanaujia SP (2021) Conserved features of the MlaD domain aid the trafficking of hydrophobic molecules. Proteins 89:1473–1488
Isom GL, Coudray N, MacRae MR, McManus CT, Ekiert DC, Bhabha G (2020) LetB structure reveals a tunnel for lipid transport across the bacterial envelope. Cell 181:653–664
Hassell AM, An G, Bledsoe RK, Bynum JM, Carter HL, Deng SJ, Gampe RT, Grisard TE, Madauss KP, Nolte RT, Rocque WJ (2007) Crystallization of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 63:72–79
Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281
Evans PR, Murshudov GN (2013) How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214
Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40:658–674
Brünger AT (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472–475
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501
Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60:2184–2195
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr D66:12–21
Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J (2000) The protein data bank and the challenge of structural genomics. Nat Struct Mol Biol 7:957–959
The UniProt Consortium (2022) UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 51:D523–D531
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Sievers F, Higgins DG (2014). In: Russell D (ed) Methods in Molecular Biology. Humana Press, Totowa
Gouet P, Robert X, Courcelle E (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL (2012) OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40:D370–D376
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Žídek A (2022) AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50:D439–D444
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Joosten RP, Te Beek TA, Krieger E, Hekkelman ML, Hooft RW, Schneider R, Sander C, Vriend G (2010) A series of PDB related databases for everyday needs. Nucleic Acids Res 39:D411–D419
Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM (2018) PDBsum: structural summaries of PDB entries. Protein Sci 27:129–134
Bond CS (2003) TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics 19:311–312
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L (2012) CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol 8:e1002708
Hagemans D, van Belzen IA, Morán Luengo T, Rüdiger SG (2015) A script to highlight hydrophobicity and charge on protein surfaces. Front Mol Biosci 2:56
Holm L (2022) Dali server: structural unification of protein families. Nucleic Acids Res 50:W210–W215
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer EL, Tosatto SC, Paladin L, Raj S, Richardson LJ, Finn RD (2021) Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419
Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49:D344–D354
Dawson NL, Lewis TE, Das S, Lees JG, Lee D, Ashford P, Orengo CA, Sillitoe I (2017) CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res 45:D289–D295
Andreeva A, Kulesha E, Gough J, Murzin AG (2020) The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res 48:D376–D382
Shukolyukov SA (2009) Aggregation of frog rhodopsin to oligomers and their dissociation to monomer: application of BN-and SDS-PAGE. Biochem (Moscow) 74:599–604
Asthana P, Singh D, Pedersen JS, Hynönen MJ, Sulu R, Murthy AV, Laitaoja M, Jänis J, Riley LW, Venkatesan R (2021) Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis. IUCrJ 8:757–774
Buslaev P, Gordeliy V, Grudinin S, Gushchin I (2016) Principal component analysis of lipid molecule conformational changes in molecular dynamics simulations. J Chem Theory Comput 12(3):1019–1028
Neumann J, Rose-Sperling D, Hellmich UA (2017) Diverse relations between ABC transporters and lipids: an overview. Biochim Biophys Acta - Biomembr 1859:605–618
Casali N, Riley LW (2007) A phylogenomic analysis of the Actinomycetales mce operons. BMC Genom 8(1):1–23
Thong S, Ercan B, Torta F, Fong ZY, Wong HY, Wenk MR, Chng SS (2016) Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5:e19042
Asthana P, Singh D, Pedersen JS, Hynönen MJ, Sulu R, Murthy AV, Laitaoja M, Jänis J, Riley LW, Venkatesan R (2021) Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis. IUCrJ 8(5):757–774
Rank L, Herring LE, Braunstein M (2021) Evidence for the mycobacterial Mce4 transporter being a multiprotein complex. J Bacteriol 203(10):10–128
Yero D, Díaz-Lobo M, Costenaro L, Conchillo-Solé O, Mayo A, Ferrer-Navarro M, Vilaseca M, Gibert I, Daura X (2021) The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm. Commun Biol 4(1):448
Schultz KM, Fischer MA, Noey EL, Klug CS (2018) Disruption of the E. Coli LptC dimerization interface and characterization of lipopolysaccharide and LptA binding to monomeric LptC. Protein Sci 27:1407–1417
Sandhya S, Rani SS, Pankaj B, Govind MK, Offmann B, Srinivasan N, Sowdhamini R (2009) Length variations amongst protein domain superfamilies and consequences on structure and function. PLoS ONE 4:e4981
Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141(2):391–396
Mao B, Pear MR, McCammon JA, Quiocho FA (1982) Hinge-bending in L-arabinose-binding protein. The Venus’s-flytrap model. J Biol Chem 257:1131–1133
Pandey S, Modak A, Phale PS, Bhaumik P (2016) High resolution structures of periplasmic glucose-binding protein of Pseudomonas putida CSV86 reveal structural basis of its substrate specificity. J Biol Chem 291:7844–7857
Chandravanshi M, Gogoi P, Kanaujia SP (2020) Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide α-glycosides through ABC transporter. FEBS J 287:1576–1597
Chandravanshi M, Samanta R, Kanaujia SP (2020) Conformational trapping of a β-Glucosides-binding protein unveils the selective two-step ligand-binding mechanism of ABC importers. J Mol Biol 432:5711–5734
Acknowledgements
The work was supported by the Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India (Grant number: ECR/2018/000013). The authors acknowledge the Central Instruments Facility (CIF) at the Indian Institute of Technology Guwahati (IITG) for providing the X-ray diffractometer (XRD). The authors are grateful to all the members of the Structural and Computational Biology laboratory (SCBL) for their continuous support. AD acknowledges the Ministry of Human Resource and Development (MHRD), Government of India, for providing the research fellowship.
Author information
Authors and Affiliations
Contributions
SPK: conceived the project, designed the study, and solved the structures; AD: performed the experiments; SPK and AD: analyzed and validated the data; SPK and AD: wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dutta, A., Kanaujia, S.P. The Structural Features of MlaD Illuminate its Unique Ligand-Transporting Mechanism and Ancestry. Protein J (2024). https://doi.org/10.1007/s10930-023-10179-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s10930-023-10179-5