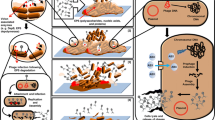
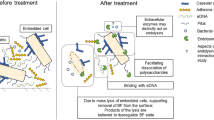
Abstract
Bacterial biofilms are widespread in the environment, and bacteria in the biofilm are highly resistant to antibiotics and possess host immune defense mechanisms, which can lead to serious clinical and environmental health problems. The increasing problem of bacterial resistance caused by the irrational use of traditional antimicrobial drugs has prompted the search for better and novel antimicrobial substances. In this paper, we review the effects of phage endolysins, modified phage endolysins, and their combination with other substances on bacterial biofilms and provide an outlook on their practical applications. Phage endolysins can specifically and efficiently hydrolyze the cell walls of bacteria, causing bacterial lysis and death. Phage endolysins have shown superior bactericidal effects in vitro and in vivo, and no direct toxicity in humans has been reported to date. The properties of phage endolysins make them promising for the prevention and treatment of bacterial infections. Meanwhile, endolysins have been genetically engineered to exert a stronger scavenging effect on biological membranes when used in combination with antibiotics and drugs. Phage endolysins are powerful weapons for controlling bacterial biofilms.
Graphical Abstract


Adapted from Maunders et al.



Similar content being viewed by others
Abbreviations
- GlcNAc:
-
N-acetylglucosamine
- MurNAc:
-
N-acetylmuramic acid
- CBD:
-
Cell wall-binding domain
- CD:
-
Catalytic domain
- EADs:
-
Enzymatically active domains
- kDa:
-
KDalton
- L-peptide:
-
Linking-peptide
- Biofilms:
-
Bacterial biofilms
References
Percival SL, Malic S, Cruz H, Williams DW (2011) Introduction to biofilms. Springer, Berlin
Habash M, Reid G (1999) Microbial biofilms: their development and significance for medical device-related infections. J Clin Pharmacol 39:887–898. https://doi.org/10.1177/00912709922008506
Maunders E, Welch M (2017) Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnx120
Simes M, Borges A, Simes L (2020) Recent trends in biofilm science and technology. Academic Press, London
Sass P, Bierbaum G (2007) Lytic activity of recombinant bacteriophage phi11 and phi12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol 73:347–352. https://doi.org/10.1128/AEM.01616-06
Monroe D (2007) Looking for chinks in the armor of bacterial biofilms. PLOS Biol 5(11):e307. https://doi.org/10.1371/journal.pbio.0050307
Loessner MJ (2005) Bacteriophage endolysins—current state of research and applications. Curr Opin Microbiol 8:480–487. https://doi.org/10.1016/j.mib.2005.06.002
Young I, Wang I-N, Roof WD (2000) Phages will out: strategies of host cell lysis. Trends Microbiol 8:120–128. https://doi.org/10.1016/s0966-842x(00)01705-4
Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R (2007) Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages φKZ and EL. Mol Microbiol 65:1334–1344. https://doi.org/10.1111/j.1365-2958.2007.05870.x
Borysowski J, Weber-Dabrowska B, Górski A (2006) Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med (Maywood) 231:366–377. https://doi.org/10.1177/153537020623100402
Navarre WW, Ton-That H, Faull KF, Schneewind O (1999) Multiple enzymatic activities of the murein hydrolase from staphylococcal phage φ11. J Biol Chem 274:15847–15856. https://doi.org/10.1074/jbc.274.22.15847
Rodríguez L, Martínez B, Zhou Y, Rodríguez A, Donovan DM, García P (2011) Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. BMC Microbiol 11:138. https://doi.org/10.1186/1471-2180-11-138
Yang H, Wang DB, Dong Q, Zhang Z, Cui Z, Deng J, Yu J, Zhang XE, Wei H (2012) Existence of separate domains in lysin PlyG for recognizing Bacillus anthracis spores and vegetative cells. Antimicrob Agents Chemother 56:5031–5039. https://doi.org/10.1128/AAC.00891-12
Young R (1992) Bacteriophage lysis: mechanism and regulation. Microbiol Rev 56:430–481. https://doi.org/10.1128/mr.56.3.430-481.1992
Vollmer W, Blanot D, de Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. https://doi.org/10.1111/j.1574-6976.2007.00094.x
Shannon R, Radford DR, Balamurugan S (2020) Impacts of food matrix on bacteriophage and endolysin antimicrobial efficacy and performance. Crit Rev Food Sci Nutr 60:1631–1640. https://doi.org/10.1080/10408398.2019.1584874
Parisien A, Allain B, Zhang J, Mandeville R, Lan CQ (2008) Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J Appl Microbiol 104:1–13. https://doi.org/10.1111/j.1365-2672.2007.03498.x
Hermoso JA, García JL, García P (2007) Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol 10:461–472. https://doi.org/10.1016/j.mib.2007.08.002
Loessner MJ, Kramer K, Ebel F, Scherer S (2002) C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol 44:335–349
Schmelcher M, Shabarova T, Eugster MR, Eichenseher F, Tchang VS, Banz M, Loessner MJ (2010) Rapid multiplex detection and differentiation of listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl Environ Microbiol 76:5745–5756. https://doi.org/10.1128/AEM.00801-10
Shen Y, Köller T, Kreikemeyer B, Nelson DC (2013) Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin. J Antimicrob Chemother 68:1818–1824. https://doi.org/10.1093/jac/dkt104
Sharma G, Rao S, Bansal A, Dang S, Gupta S, Gabrani R (2014) Pseudomonas aeruginosa biofilm: potential therapeutic targets. Biologicals 42:1–7. https://doi.org/10.1016/j.biologicals.2013.11.001
Indiani C, Sauve K, Raz A, Abdelhady W, Xiong YQ, Cassino C, Bayer AS, Schuch R (2019) The antistaphylococcal Lysin, CF-301, activates key host factors in human blood to potentiate methicillin-resistant Staphylococcus aureus bacteriolysis. Antimicrob Agents Chemother 63:e02291-e2318. https://doi.org/10.1128/AAC.02291-18
Poonacha N, Nair S, Desai S, Tuppad D, Hiremath D, Mohan T, Vipra A, Sharma U (2017) Efficient killing of planktonic and biofilm-embedded coagulase-negative Staphylococci by bactericidal protein P128. Antimicrob Agents Chemother 61:e00457-e517. https://doi.org/10.1128/AAC.00457-17
Singh PK, Donovan DM, Kumar A (2014) Intravitreal injection of the chimeric phage endolysin Ply187 protects mice from Staphylococcus aureus endophthalmitis. Antimicrob Agents Chemother 58:4621–4629. https://doi.org/10.1128/AAC.00126-14
Lood R, Winer BY, Pelzek AJ, Diez-Martinez R, Thandar M, Euler CW, Schuch R, Fischetti VA (2015) Novel phage lysin capable of killing the multidrug-resistant Gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother 59:1983–1991. https://doi.org/10.1128/AAC.04641-14
Wang Y, Sun JH, Lu CP (2009) Purified recombinant phage lysin LySMP: an extensive spectrum of lytic activity for swine streptococci. Curr Microbiol 58(6):609–615. https://doi.org/10.1007/s00284-009-9379-x
Fang, Y. Z., Wang, Y., & Sun, J. H. (2011) Expression of the phage lysin lysmp in Lactococcus lactis and studies on its antibiotic bioactivities. Journal of Shanghai Jiaotong University (Agricultural Science)
Leitch EC, Willcox MD (1999) Lactoferrin increases the susceptibility of S. epidermidis biofilms to lysozyme and vancomycin. Curr Eye Res 19:12–19. https://doi.org/10.1076/ceyr.19.1.12.5342
Hukić M, Seljmo D, Ramovic A, Ibrišimović MA, Dogan S, Hukic J, Bojic EF (2018) The effect of lysozyme on reducing biofilms by Staphylococcus aureus, Pseudomonas aeruginosa, and Gardnerella vaginalis: an in vitro examination. Microb Drug Resist 24:353–358. https://doi.org/10.1089/mdr.2016.0303
Ellison R, Giehl T, Laforce FM (1988) Damage of the outer membrane of enteric Gram-negative bacteria by lactoferrin and transferring. Infect Immun 56(11):2774–2781. https://doi.org/10.1128/IAI.56.11.2774-2781.1988
Lee CK, Rubin LG, Moldwin RM (1995) Synergy between protamine and vancomycin in the treatment of Staphylococcus epidermidis biofilms. Urology 45:720–724. https://doi.org/10.1016/S0090-4295(99)80074-0
Teichman JM (1994) Protamine sulphate and vancomycin are synergistic against Staphylococcus epidermidis prosthesis infections in vivo. J Urol 152:213–216. https://doi.org/10.1016/S0022-5347(17)32864-1
Yeaman MR, Norman DC, Bayer AS (1992) Platelet microbicidal protein enhances antibiotic–induced killing of and postantibiotic effect in Staphylococcus aureus. Antimicrob Agents Chemother 36:1665–1670. https://doi.org/10.1128/AAC.36.8.1665
Kuiper Jesse WP, Hogervorst Jolanda MA, Herpers Bjorn L, Bakker Astrid D, Jenneke KN, Nolte Peter A, Krom Bastiaan P (2021) The novel endolysin XZ700 effectively treats MRSA biofilms in two biofilm models without showing toxicity on human bone cells. Biofouling. https://doi.org/10.1080/08927014.2021.1887151
Żebrowska J, Żołnierkiewicz O, Ponikowska M, Puchalski M, Krawczun N, Makowska J, Skowron P (2022) Cloning and characterization of a thermostable endolysin of bacteriophage TP-84 as a potential disinfectant and biofilm-removing biological agent. Int J Mol Sci 23(14):7612. https://doi.org/10.3390/ijms23147612
Christine L, Vera O, Lenka PT, Timo S, Rocío B, Leen VS, Mario V, Lorenzo C (2022) Preclinical data on the gardnerella-specific endolysin PM-477 indicate its potential to improve the treatment of bacterial vaginosis through enhanced biofilm removal and avoidance of resistance. Antimicrob Agents Chemother. https://doi.org/10.1128/aac.02319-21
William J, Alicia W, Frederique KW, Christopher D, Lee BJ, Suzanne H, David C, Matthew C, Gordon R, Ryan K (2023) In vitro bacterial vaginosis biofilm community manipulation using endolysin therapy. Biofilm. https://doi.org/10.1016/j.bioflm.2022.100101
Fursov MV, Abdrakhmanova RO, Antonova NP, Vasina DV, Kolchanova AD, Bashkina OA, Rubalsky OV, Samotrueva MA, Potapov VD, Makarov VV, Yudin SM, Gintsburg AL, Tkachuk AP, Gushchin VA, Rubalskii EO (2020) Antibiofilm activity of a broad-range recombinant endolysin LysECD7. Vitro In Vivo Study. https://doi.org/10.3390/v12050545
Hou-Qi N, Hong L, Jing-Xue W (2021) Synergistic effects of endolysin Lysqdvp001 and ε-poly-lysine in controlling Vibrio parahaemolyticus and its biofilms. Int J Food Microbiol. https://doi.org/10.1016/j.ijfoodmicro.2021.109112
Baliga P, Goolappa PT, Shekar M, Kallappa GS (2022) Cloning, characterization, and antibacterial properties of endolysin LysE against planktonic cells and biofilms of Aeromonas hydrophila. Probiotics Antimicrob Proteins. https://doi.org/10.1007/s12602-021-09880-7
Oh HK, Hwang YJ, Hong HW, Myung H (2021) Comparison of Enterococcus faecalis biofilm removal efficiency among bacteriophage PBEF129, Its endolysin, and cefotaxime. Viruses 13:426. https://doi.org/10.3390/v13030426
Oliveira H, Thiagarajan V, Walmagh M, Sillankorva S, Azeredo J (2014) A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against Gram-Negative pathogens in presence of weak acids. PLoS ONE 9(10):1–11. https://doi.org/10.1371/journal.pone.0108376
Zhang J, Xu L-L, Gan Dan, Zhang X (2018) In vitro study of bacteriophage AB3 endolysin LysAB3 activity against Acinetobacter baumannii biofilm and biofilm-bound A. baumannii. Clin Lab 64:6. https://doi.org/10.7754/Clin.Lab.2018.180342
Fenton M, Ross P, McAuliffe O, O’Mahony J, Coffey A (2010) Recombinant bacteriophage lysins as antibacterials. Bioeng Bugs 1:9–16. https://doi.org/10.4161/bbug.1.1.9818
Lu TK, Collins JJ (2007) Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci USA 104:11197–11202. https://doi.org/10.1073/pnas.0704624104
Pritchard DG, Dong S, Kirk MC, Cartee RT, Baker JR (2007) LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl Environ Microbiol 73:7150–7154. https://doi.org/10.1128/AEM.01783-07
Donovan DM, Dong S, Garrett W, Rousseau GM, Moineau S, Pritchard DG (2006) Peptidoglycan hydrolase fusions maintain their parental specificities. Appl Environ Microbiol 72:2988–2996. https://doi.org/10.1128/AEM.72.4.2988-2996.2006
Becker SC, Foster-Frey J, Stodola AJ, Anacker D, Donovan DM (2009) Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 443:32–41. https://doi.org/10.1016/j.gene.2009.04.023
Rodríguez-Rubio L, Martínez B, Rodríguez A, Donovan DM, García P (2012) Enhanced staphylolytic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88 HydH5 virion-associated peptidoglycan hydrolase: fusions, deletions, and synergy with LysH5. Appl Environ Microbiol 78:2241–2248. https://doi.org/10.1128/AEM.07621-11
Jagielska E, Chojnacka O, Sabała I (2016) LytM fusion with SH3b-like domain expands its activity to physiological conditions. Microb Drug Resist 22:461–469. https://doi.org/10.1089/mdr.2016.0053
Yang H, Zhang Y, Yu J, Huang Y, Zhang XE, Wei H (2014) Novel chimeric lysin with high-level antimicrobial activity against methicillin-resistant Staphylococcus aureus in vitro and in vivo. Antimicrob Agents Chemother 58:536–542. https://doi.org/10.1128/AAC.01793-13
Dong Q, Wang J, Yang H, Wei C, Yu J, Zhang Y, Huang Y, Zhang XE, Wei H (2015) Construction of a chimeric lysin Ply187N-V12C with extended lytic activity against staphylococci and streptococci. Microb Biotechnol 8:210–220. https://doi.org/10.1111/1751-7915.12166
Yang H, Zhang Y, Yu J, Huang Y, Xian-En H (2014) Novel chimeric lysin with high-level antimicrobial activity against methicillin-resistant Staphylococcus aureus in vitro and in vivo. Antimicrob Agents Chemother 58:536–542. https://doi.org/10.1128/AAC.01793-13
Yang H, Zhang Y, Huang Y, Yu J, Wei H (2014) Degradation of methicillin-resistant Staphylococcus aureus biofilms using a chimeric lysin. Biofouling 30:667–674. https://doi.org/10.1080/08927014.2014.905927
Fernandes S, Proença D, Cantante C, Silva FA, Leandro C, Lourenço S, Milheiriço C, de Lencastre H, Cavaco-Silva P, Pimentel M, são-José C, (2012) Novel chimerical endolysins with broad antimicrobial activity against methicillin-resistant Staphylococcus aureus. Microb Drug Resist 18:333–343. https://doi.org/10.1089/mdr.2012.0025
Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA (2011) A novel chimeric lysin shows superiority to Mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother 55:738–744. https://doi.org/10.1128/AAC.00890-10
Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA (2010) Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:1603–1612. https://doi.org/10.1128/AAC.01625-09
Signph PK, Donovan DM, Kumar A (2014) Intravitreal injection of the chimeric phage endolysin Ply187 protects mice from Staphylococcus aureus endophthalmitis. Antimicrob Agents Chemother 58(8):4621–4629. https://doi.org/10.1128/AAC.00126-14
Schmelcher M, Tchang VS, Loessner MJ (2011) Domain shuffling and module engineering of Listeria phage endolysins for enhanced lytic activity and binding affinity. Microb Biotechnol 4:651–662. https://doi.org/10.1111/j.1751-7915.2011.00263.x
Vasala A, Välkkilä M, Caldentey J, Alatossava T (1995) Genetic and biochemical characterization of the Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H lysin. Appl Environ Microbiol 61:4004–4011. https://doi.org/10.1128/aem.61.11.4004-4011.1995
Loessner MJ, Gaeng S, Scherer S (1999) Evidence for a holin-like protein gene fully embedded out of frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J Bacteriol 181:4452–4460. https://doi.org/10.1128/JB.181.15.4452-4460.1999
Meng WU, Hai-Rong LU, Qingshan H (2016) Expression of CHAP structural domain of Staphylococcus aureus phage lytic enzyme Ply187 and analysis of antibacterial activity. Biotechnol Bull 32(9):232–238. https://doi.org/10.13560/j.cnki.biotech.bull.1985.2016.09.031
Horgan M, O’Flynn G, Garry J, Cooney J, Coffey A, Fitzgerald GF, Ross RP, McAuliffe O (2009) Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl Environ Microbiol 75:872–874. https://doi.org/10.1128/AEM.01831-08
Fenton M, Ross RP, McAuliffe O, O’Mahony J, Coffey A (2011) Characterization of the staphylococcal bacteriophage lysin CHAP(K). J Appl Microbiol 111:1025–1035. https://doi.org/10.1111/j.1365-2672.2011.05119.x
Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R (2011) Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J Bacteriol 193:5477–5486. https://doi.org/10.1128/JB.00439-11
Fenton M, Keary R, McAuliffe O, Ross RP, O’Mahony J, Coffey A (2013) Bacteriophage-derived peptidase CHAP(K) eliminates and prevents staphylococcal biofilms. Int J Microbiol 2013:625341. https://doi.org/10.1155/2013/625341CHAP(K)
Violeta RC, Pedro G, Gema DP, Ernesto G, Matilde G, Lorena H et al (2007) In vitro interactions of lyta, the major pneumococcal autolysin, with two bacteriophage lytic enzymes (cpl-1 and pal), cefotaxime and moxifloxacin against antibiotic-susceptible and -resistant Streptococcus pneumoniae strains. J Antimicrob Chemother 5:1159–62
Filatova LY, Donovan DM, Ishnazarova NT, Foster-Frey JA, Becker SC, Pugachev VG, Balabushevich NG, Dmitrieva NF, Klyachko NL (2016) A chimeric LysK-lysostaphin fusion enzyme lysing Staphylococcus aureus cells: a study of both kinetics of inactivation and specifics of interaction with anionic polymers. Appl Biochem Biotechnol 180:544–557. https://doi.org/10.1007/s12010-016-2115-7
Djurkovic S, Loeffler JM, Fischetti VA (2005) Synergistic killing of Streptococcus pneumoniae with the bacteriophage lytic enzyme cpl-1 and penicillin or gentamicin depends on the level of penicillin resistance. Antimicrob Agents Chemother 49:1225–1228. https://doi.org/10.1128/AAC.49.3.1225-1228.2005
McCarthy MW (2022) Exebacase: a novel approach to the treatment of Staphylococcal infections. Drugs R D 22:113–117. https://doi.org/10.1007/s40268-022-00383-6
Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay JP, Miller S, Volckaert G, Lavigne R (2014) Engineered endolysin-based “artilysins” to combat multidrug-resistant Gram-negative pathogens. mBio 5:e01379–e01314
Lim JA, Shin H, Kang DH, Ryu S (2012) Characterization of endolysin from a Salmonella typhimurium-infecting bacteriophage SPN1S. Res Microbiol 163:233–241. https://doi.org/10.1016/j.resmic.2012.01.002
Plotka M, Kapusta M, Dorawa S, Kaczorowska AK, Kaczorowski TTS (2019) Ts2631 endolysin from the extremophilic Thermus scotoductus bacteriophage vB_Tsc2631 as an antimicrobial agent against Gram-negative multidrug-resistant bacteria. Viruses 11:657. https://doi.org/10.3390/v11070657
Liu A, Wang Y, Cai X, Jiang S, Cai X, Shen L, Liu Y, Han G, Chen S, Wang J, Wu W, Li C, Liu S, Wang X (2019) Characterization of endolysins from bacteriophage LPST10 and evaluation of their potential for controlling Salmonella Typhimurium on lettuce. LWT 114:108372. https://doi.org/10.1016/j.lwt.2019.108372
Berini F, Orlandi V, Gornati R, Bernardini G, Marinelli F (2022) Nanoantibiotics to fight multidrug resistant infections by Gram-positive bacteria: hope or reality? Biotechnol Adv 57:107948. https://doi.org/10.1016/j.biotechadv.2022.107948
Liu Y, Shi L, Su L, van der Mei HC, Jutte PC, Ren Y, Busscher HJ (2019) Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control. Chem Soc Rev 48:428–446. https://doi.org/10.1039/c7cs00807d
Liu G, Xu Y, Han Y, Wu J, Xu J, Meng H, Zhang X (2017) Immobilization of lysozyme proteins on a hierarchical zeolitic imidazolate framework (ZIF-8). Dalton Trans 46:2114–2121. https://doi.org/10.1039/c6dt04582k
Wang Y, Li S, Jin M, Han Q, Liu S, Chen X, Han Y (2020) Enhancing the thermo-stability and anti-bacterium activity of lysozyme by immobilization on chitosan nanoparticles. Int J Mol Sci 21:1635. https://doi.org/10.3390/ijms21051635
Liu Y, Sun Y, Xu Y, Feng H, Fu S, Tang J, Liu W, Sun D, Jiang H, Xu S (2013) Preparation and evaluation of lysozyme-loaded nanoparticles coated with poly-γ-glutamic acid and chitosan. Int J Biol Macromol 59:201–207. https://doi.org/10.1016/j.ijbiomac.2013.04.065
Chhibber S, Nag D, Bansal S (2013) Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and abacteriophage. BMC Microbiol 13:174. https://doi.org/10.1186/1471-2180-13-174
Zhang Y, Hu Z (2012) Combined treatment of Pseudomonas aeruginosa biofilms with bacteriophages and chlorine. Biotechnol Bioeng 110(1):286–295. https://doi.org/10.1002/bit.24630
Kovalskaya N, Foster-Frey J, Donovan DM (2016) Antimicrobial activity of bacteriophage endolysin produced in Nicotiana benthamiana plants. J Microbiol Biotechnol 26(1):160–170. https://doi.org/10.4014/jmb.1505.05060
Nelson DC, Schmelcher M, Rodriguez-Rubio L, Klumpp J, Pritchard DG, Dong S, Donovan DM (2012) Chapter 7—endolysins as antimicrobials. Adv Virus Res 83:299–365. https://doi.org/10.1016/B978-0-12-394438-2.00007-4
Acknowledgements
The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The authors declare no conflict of interest.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities, grant number E1E40506 and Weiqiao-UCAS Special Projects on Low-Carbon Technology Development (No. GYY-DTFZ-2022-008).
Author information
Authors and Affiliations
Contributions
Conceptualization, BL, and XL; methodology, BL; software, ZL; validation, QG; formal analysis, BL; investigation, XL; resources, XL; data curation, XG; writing—original draft preparation, BL; writing—review and editing, QG; visualization, ZL, and BL supervision, XL; project administration, XL; funding acquisition, XL. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, B., Guo, Q., Li, Z. et al. Bacteriophage Endolysin: A Powerful Weapon to Control Bacterial Biofilms. Protein J 42, 463–476 (2023). https://doi.org/10.1007/s10930-023-10139-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10139-z