Abstract
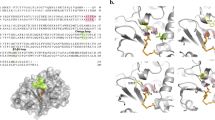
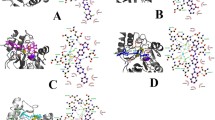
The rise of New Delhi metallo beta-lactamase (NDM) producing bacteria imposes a significant threat to the treatment of bacterial infections due to their broad spectrum against beta-lactams. The activity of metallo beta-lactamases is affected by active site residues as well as residues near the active site. Therefore, we aimed to identify the amino acid residues around the active site of NDM-4 which influence its function. To achieve that, seven substitution mutations (S191A, D192A, S213A, K216A, S217A, D223A and D225A) of NDM-4 were generated through site-directed mutagenesis. Out of these, expression of NDM-4_D192A and NDM-4_S217A in Escherichia coli cells increased the beta-lactam susceptibility as compared to NDM-4. Further, proteins were purified to assess the effect of substitution mutations on zinc content, in vitro catalytic efficiency, and stability of NDM-4. The catalytic efficiency was reduced for these mutants (D192A and S217A) towards beta-lactam substrates, while the thermal stability remained insubstantial as compared to NDM-4. However, the purified NDM-4_D192A exhibited altered zinc content. In silico studies reveal that these changes might be the outcomes of alterations in hydrogen bonding networks and substrate interactions. Taken together, we infer that the D192 and the S217 residues play a substantial role in the activity of NDM-4.





Similar content being viewed by others
References
Hall BG, Barlow M (2005) Revised Ambler classification of β-lactamases. J Antimicrob Chemother 55(6):1050–1051
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR (2009) Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53(12):5046–5054
Rolain J, Parola P, Cornaglia G (2010) New Delhi metallo-beta-lactamase (NDM-1): towards a new pandemia? Clin Microbiol Infect 16(12):1699–1701
Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z (2019) NDM metallo-β-lactamases and their bacterial producers in health care settings.Clinical microbiology reviews32(2)
Bush K (2010) Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol 13(5):558–564
Jing-yi Z, Yuan-qi Z, Yan-nian L, Xiao-dong M, Li-ping Y, Cha X, Qin P, Jin-long M (2015) Coexistence of SFO-1 and NDM-1 β-lactamase genes and fosfomycin resistance gene fosA3 in an Escherichia coli clinical isolate. FEMS Microbiol Lett 362(1):1–7
Karthikeyan K, Thirunarayan M, Krishnan P (2010) Coexistence of bla OXA-23 with bla NDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother 65(10):2253–2254
King D, Strynadka N (2011) Crystal structure of New Delhi metallo-β‐lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20(9):1484–1491
Marcoccia F, Bottoni C, Sabatini A, Colapietro M, Mercuri PS, Galleni M, Kerff F, Matagne A, Celenza G, Amicosante G (2016) Kinetic study of laboratory mutants of NDM-1 metallo-β-lactamase and the importance of an isoleucine at position 35. Antimicrob Agents Chemother 60(4):2366–2372
Linciano P, Cendron L, Gianquinto E, Spyrakis F, Tondi D (2018) Ten years with New Delhi metallo-β-lactamase-1 (NDM-1): from structural insights to inhibitor design. ACS Infect Dis 5(1):9–34
Biedenbach D, Bouchillon S, Hackel M, Hoban D, Kazmierczak K, Hawser S, Badal R (2015) Dissemination of NDM metallo-β-lactamase genes among clinical isolates of Enterobacteriaceae collected during the SMART global surveillance study from 2008 to 2012. Antimicrob Agents Chemother 59(2):826–830
Nordmann P, Boulanger AE, Poirel L (2012) NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother 56(4):2184–2186
Bahr G, Vitor-Horen L, Bethel CR, Bonomo RA, González LJ, Vila AJ (2018) Clinical evolution of New Delhi metallo-β-lactamase (NDM) optimizes resistance under Zn (II) deprivation. Antimicrob Agents Chemother 62(1):e01849–e01817
Stewart AC, Bethel CR, VanPelt J, Bergstrom A, Cheng Z, Miller CG, Williams C, Poth R, Morris M, Lahey O (2017) Clinical variants of New Delhi metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect Dis 3(12):927–940
Chiou J, Leung TY-C, Chen S (2014) Molecular mechanisms of substrate recognition and specificity of New Delhi metallo-β-lactamase. Antimicrob Agents Chemother 58(9):5372–5378
Chen J, Chen H, Shi Y, Hu F, Lao X, Gao X, Zheng H, Yao W (2013) Probing the effect of the non-active-site mutation Y229W in New Delhi metallo-β-lactamase-1 by site-directed mutagenesis, kinetic studies, and molecular dynamics simulations.PLoS One8(12)
Kumar G, Issa B, Kar D, Biswal S, Ghosh AS (2017) E152A substitution drastically affects NDM-5 activity.FEMS Microbiology Letters364(3)
Kumar G, Issa B, Biswal S, Jain D, Bhattacharjee A, Ghosh AS (2020) Glutamic acid at position 152 and serine at position 191 are key residues required for the metallo-β-lactamase activity of NDM-7. Int J Antimicrob Agents 55(1):105824
Ali A, Azam MW, Khan AU (2018) Non-active site mutation (Q123A) in New Delhi metallo-β-lactamase (NDM-1) enhanced its enzyme activity. 112:1272–1277
Choudhury NA, Paul D, Chakravarty A, Bhattacharjee A, Chanda DD (2018) IncX3 plasmid mediated occurrence of blaNDM-4 within Escherichia coli ST448 from India. J Infect Public Health 11(1):111–114
Guzman L-M, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177(14):4121–4130
Bansal A, Kar D, Murugan RA, Mallick S, Dutta M, Pandey SD, Chowdhury C, Ghosh AS (2015) A putative low-molecular-mass penicillin-binding protein (PBP) of Mycobacterium smegmatis exhibits prominent physiological characteristics of DD-carboxypeptidase and beta-lactamase. Microbiology 161(5):1081–1091
Kumar G, Biswal S, Nathan S, Ghosh AS (2018) Glutamate residues at positions 162 and 164 influence the beta-lactamase activity of SHV-14 obtained from Klebsiella pneumoniae. FEMS Microbiol Lett 365(2):fnx259
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
CLSI C (2016) Performance standards for antimicrobial susceptibility testing. Clinical Lab Standards Institute
Fast W, Wang Z, Benkovic SJ (2001) Familial mutations and zinc stoichiometry determine the rate-limiting step of nitrocefin hydrolysis by metallo-β-lactamase from Bacteroides fragilis. Biochemistry 40(6):1640–1650
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7(1):539
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291
Tian W, Chen C, Lei X, Zhao J, Liang J (2018) CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 46(W1):W363–W367
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Bekker H, Berendsen H, Dijkstra E, Achterop S, Vondrumen R, VANDERSPOEL D, Sijbers A, Keegstra H, Renardus M (1993) Gromacs-a parallel computer for molecular-dynamics simulations. In: 4th International Conference on Computational Physics (PC 92). World Scientific Publishing, pp 252–256
Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E (2021) gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput 17(10):6281–6291
Kumar G, Issa B, Biswal S, Jain D, Bhattacharjee A, Ghosh ASJIjoaa (2020) Glutamic acid at position 152 and serine at position 191 are key residues required for the metallo-β-lactamase activity of NDM-7. 55(1):105824
Ali A, Kumar R, Iquebal MA, Jaiswal S, Kumar D, Khan AU (2019) The role of conserved residues in the catalytic activity of NDM-1: an approach involving site directed mutagenesis and molecular dynamics. Phys Chem Chem Phys 21(32):17821–17835
Sun Z, Hu L, Sankaran B, Prasad B, Palzkill T (2018) Differential active site requirements for NDM-1 β-lactamase hydrolysis of carbapenem versus penicillin and cephalosporin antibiotics. Nat Commun 9(1):1–14
Zhang H, Ma G, Zhu Y, Zeng L, Ahmad A, Wang C, Pang B, Fang H, Zhao L, Hao Q (2018) Active-site conformational fluctuations promote the enzymatic activity of NDM-1. Antimicrob Agents Chemother 62(11):e01579–e01518
Thapa A, Upreti MK, Bimali NK, Shrestha B, Sah AK, Nepal K, Dhungel B, Adhikari S, Adhikari N, Lekhak B (2022) Detection of NDM Variants (bla NDM-1, bla NDM-2, bla NDM-3) from Carbapenem-Resistant Escherichia coli and Klebsiella pneumoniae: First Report from Nepal. Infect Drug Resistance:4419–4434
Chen J, Chen H, Zhu T, Zhou D, Zhang F, Lao X, Zheng H (2014) Asp120Asn mutation impairs the catalytic activity of NDM-1 metallo-β-lactamase: experimental and computational study. Phys Chem Chem Phys 16(14):6709–6716
Ali A, Gupta D, Khan AU (2021) Role of non-active site residues in maintaining New Delhi metallo-β-lactamase-1 (NDM-1) function: an approach of site-directed mutagenesis and docking. FEMS Microbiol Lett 368(4):fnz003
González LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ (2016) Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat Chem Biol 12(7):516
Acknowledgements
The authors are highly thankful to Prof. Amitabha Bhattacharjee, Department of Microbiology, Assam University, Silchar, India for providing blaNDM−4 carrying E. coli ST44.
Funding
This work was funded partially by two grants, one from the Department of Biotechnology (DBT), Government of India [grant # BT/PR24255/NER/95/716/2017] and another from Council of Scientific and Industrial Research (CSIR), Govt. of India [grant # 27(0367)/20/EMR-II ].
Author information
Authors and Affiliations
Contributions
Jyoti Verma: Performed experiment and wrote the first draft; Diamond Jain: Performed experiment, edited the manuscript, prepared the revised manuscript; Aditya Prasad Panda: Bioinformatics analysis and relevant writing; Shri Kant: Bioinformatics analysis and relevant writing; Gaurav Kumar: provided relevant suggestions for improvement; Anindya S Ghosh: Conceptualization, supervision and preparation of the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
None to declare.
Ethical approval
Not required.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Verma, J., Jain, D., Panda, A.P. et al. Involvement of the non-active site Residues in the Catalytic Activity of NDM-4 Metallo beta-lactamase. Protein J 42, 316–326 (2023). https://doi.org/10.1007/s10930-023-10124-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10124-6