Abstract
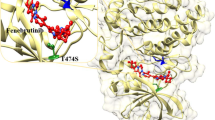
B-cell lymphoma 2 (Bcl2) is a key protein regulator of apoptosis. The hydrophobic groove in Bcl2 is a unique structural feature to this class of enzymes and found to have a profound impact on protein overall structure, function, and dynamics. Dynamics of the hydrophobic groove is an essential determinant of the catalytic activity of Bcl2, an implicated protein in Waldenström macroglobulinemia (WM). The mobility of α3–α4 helices around the catalytic site of the protein remains crucial to its activity. The preferential binding mechanisms of the multi-catalytic sites of the Bcl2 enzyme have been a subject of debate in the literature. In addition to our previous report on the same protein, herein, we further investigate the preferential binding modes and the conformational implications of Venetoclax-JQ1 dual drug binding at both catalytic active sites of Bcl2. Structural analysis revealed asymmetric α3–α4 helices movement with the expansion of the distance between the α3 and α4 helix in Venetoclax-JQ1 dual inhibition by 15.2% and 26.3%, respectively when compared to JQ1 and Venetoclax individual drug inhibition—resulting in remarkable widening of hydrophobic groove. Moreso, a reciprocal enhanced binding effect was observed: Venetoclax increased the binding affinity of JQ1 by 11.5%, while the JQ1 fostered the binding affinity of Venetoclax by 16.3% compared with individual inhibition of each drug. This divergence has also resulted in higher protein stability, and prominent correlated motions were observed with the least fluctuations and multiple van der Waals interactions. Findings offer vital conformational dynamics and structural mechanisms of enzyme-single ligand and enzyme-dual ligand interactions, which could potentially shift the current therapeutic protocol of Waldenström macroglobulinemia.













Similar content being viewed by others
References
Swerdlow S, Cook J, Sohani A et al (2017) L lymphoma. In: Swerdlow S, Campo E, Harris NE (eds) WHO classification of tumour of hematopoietic and lymphoid tissues. IARC Press, Lyon, pp 232–235
Owen RG, Treon SP, Al-Katib A et al (2003) Clinicopathological definition of Waldenstrom’s macroglobulinemia: consensus panel recommendations from the second international workshop on Waldenstrom’s macroglobulinemia. Semin Oncol 30(2):110–115
Buske C, Sadullah S, Kastritis E et al (2018) Treatment and outcome patterns in European patients with Waldenström’s macroglobulinaemia: a large, observational, retrospective chart review. Lancet Haematol 5(7):e299–e309
Olszewski AJ, Treon SP, Castillo JJ (2016) Evolution of management and outcomes in waldenström macroglobulinemia: a population-based analysis. Oncologist 21(11):1377–1386. https://doi.org/10.1634/theoncologist.2016-0126
Treon SP, Ioakimidis L, Soumerai JD et al (2009) Primary therapy of waldenström macroglobulinemia with bortezomib, dexamethasone, and rituximab: WMCTG clinical trial 05–180. J Clin Oncol 27(23):3830–3835. https://doi.org/10.1200/JCO.2008.20.4677
Dimopoulos MA, Anagnostopoulos A, Kyrtsonis M-C et al (2007) Primary treatment of waldenström macroglobulinemia with dexamethasone, rituximab, and cyclophosphamide. J Clin Oncol 25(22):3344–3349. https://doi.org/10.1200/JCO.2007.10.9926
Rummel MJ, Niederle N, Maschmeyer G et al (2013) Bendamustine plus rituximab versus CHOP plus rituximab as first-line treatment for patients with indolent and mantle-cell lymphomas: an open-label, multicentre, randomised, phase 3 non-inferiority trial. Lancet 381(9873):1203–1210
Dimopoulos MA, Kastritis E (2019) How i treat waldenström macroglobulinemia. Blood 134(23):2022–2035
Matissek SJ, Han W, Karbalivand M et al (2021) Epigenetic targeting of waldenström macroglobulinemia cells with BET inhibitors synergizes with BCL2 or histone deacetylase inhibition. Epigenomics 13(2):129–144. https://doi.org/10.2217/epi-2020-0189
Yip KW, Reed JC (2008) Bcl-2 family proteins and cancer. Oncogene 27(50):6398–6406
Tsujimoto Y, Finger L, Yunis J, Nowell P, Croce C (1984) Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 226(4678):1097–1099. https://doi.org/10.1126/science.6093263
Tsujimoto Y, Cossman J, Jaffe E, Croce C (1985) Involvement of the bcl-2 gene in human follicular lymphoma. Science 228(4706):1440–1443. https://doi.org/10.1126/science.3874430
Tsujimoto Y (1989) Stress-resistance conferred by high level of bcl-2 alpha protein in human B lymphoblastoid cell. Oncogene 4(11):1331–1336
Petros AM, Medek A, Nettesheim DG et al (2001) Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci 98(6):3012–3017. https://doi.org/10.1073/pnas.041619798
Yin X-M, Oltvai ZN, Korsmeyer SJ (1994) BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature 369(6478):321–323
Ku B, Liang C, Jung JU, Oh B-H (2011) Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res 21(4):627–641
Shahbazi J, Liu PY, Atmadibrata B et al (2016) The bromodomain inhibitor JQ1 and the histone deacetylase inhibitor panobinostat synergistically reduce N-myc expression and induce anticancer effects. Clin Cancer Res 22(10):2534–2544. https://doi.org/10.1158/1078-0432.CCR-15-1666
Delbridge ARD, Strasser A (2015) The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ 22(7):1071–1080
Besbes S, Mirshahi M, Pocard M, Billard C (2015) New dimension in therapeutic targeting of BCL-2 family proteins. Oncotarget 6(15):12862–12871
Cory S, Roberts AW, Colman PM, Adams JM (2016) Targeting BCL-2-like proteins to kill cancer cells. Trends Cancer 2(8):443–460
Correia C, Lee SH, Meng XW et al (2015) Emerging understanding of Bcl-2 biology: Implications for neoplastic progression and treatment. Biochim Biophys Acta Mol Cell Res 1853(7):1658–1671
Touré BB, Miller-Moslin K, Yusuff N et al (2013) The Role of the acidity of N-heteroaryl sulfonamides as inhibitors of Bcl-2 family protein-protein interactions. ACS Med Chem Lett 4(2):186–190
Oltersdorf T, Elmore SW, Shoemaker AR et al (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435(7042):677–681
Tse C, Shoemaker AR, Adickes J et al (2008) ABT-263: A potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 68(9):3421–3428
Lee EF, Fairlie WD (2019) The structural biology of Bcl-xL. Int J Mol Sci 20(9):2234
Berman HM, Battistuz T, Bhat TN et al (2002) The protein data bank. Acta Crystallogr D 58(1):899–907
Birkinshaw RW, Gong J, Luo CS et al (2019) Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations. Nat Commun 10(1):2385
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Kusumaningrum S, Budianto E, Kosela S, Sumaryono W, Juniarti F (2014) The molecular docking of 1,4-naphthoquinone derivatives as inhibitors of Polo-like kinase 1 using Molegro Virtual Docker. J. Appl Pharm Sci 4:47–53
Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49(11):3315–3321
Allouche A (2012) Software news and updates gabedit—a graphical user interface for computational chemistry softwares. J Comput Chem 32:174–182
Trott O, Olson A (2010) Autodock vina: improving the speed and accuracy of docking. J Comput Chem 31(2):455–461
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234
Mhlongo NN, Ebrahim M, Skelton AA, Kruger HG, Williams IH, Soliman MES (2015) Dynamics of the thumb-finger regions in a GH11 xylanase Bacillus circulans: comparison between the Michaelis and covalent intermediate. RSC Adv 5(100):82381–82394
Ramharack P, Oguntade S, Soliman MES (2017) Delving into Zika virus structural dynamics-a closer look at NS3 helicase loop flexibility and its role in drug discovery. RSC Adv 7(36):22133–22144
Case DA, Walker RC, Cheatham TE et al (2018) Amber. Univ Calif San Fr 18:1–923
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174. https://doi.org/10.1002/jcc.20035
Grest GS, Kremer K (1986) Molecular dynamics simulation for polymers in the presence of a heat bath. Phys Rev A 33(5):3628–3631. https://doi.org/10.1103/PhysRevA.33.3628
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81(8):3684–3690. https://doi.org/10.1063/1.448118
Ryckaert J-P, Ciccotti G, Berendsen HJ (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341
Roe DR, Cheatham TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9(7):3084–3095. https://doi.org/10.1021/ct400341p
Seifert E (2014) OriginPro 9.1: scientific data analysis and graphing software—software review. J Chem Inf Model 54(5):1552
Abdullahi M, Olotu FA, Soliman ME (2017) Dynamics of allosteric modulation of lymphocyte function associated antigen-1 closure-open switch: unveiling the structural mechanisms associated with outside-in signaling activation. Biotechnol Lett 39(12):1843–1851
Olotu FA, Soliman MES (2018) From mutational inactivation to aberrant gain-of-function: Unraveling the structural basis of mutant p53 oncogenic transition. J Cell Biochem 119(3):2646–2652
Wan H, Hu JP, Tian XH, Chang S (2013) Molecular dynamics simulations of wild type and mutants of human complement receptor 2 complexed with C3d. Phys Chem Chem Phys 15(4):1241–1251
Chang S, Hu JP, Lin PY, Jiao X, Tian XH (2010) Substrate recognition and transport behavior analyses of amino acid antiporter with coarse-grained models. Mol Biosyst 6(12):2430–2438
Fakhar Z, Govender T, Maguire GEM et al (2017) Differential flap dynamics in transpeptidase2 from mycobacterium tuberculosis revealed by molecular dynamics. Mol Biosyst 13(6):1223–1234
David CC, Jacobs DJ (2014) Principal component analysis: a method for determining the essential dynamics of proteins. Methods Mol Biol 14:193–226
Levy RM, Srinivasan AR, Olson WK, McCammon JA (1984) Quasi-harmonic method for studying very low frequency modes in proteins. Biopolymers 23(6):1099–1112
Chen J, Wang J, Zhu W (2014) Binding modes of three inhibitors 8CA, F8A and I4A to A-FABP studied based on molecular dynamics simulation. PLoS ONE 9(6):e99862
Ichiye T, Karplus M (1991) Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins Struct Funct Genet 11(3):205–217
Humphrey W, Dalke A, Schulten KVMD (1996) Visual molecular dynamics. J Mol Graph 14(1):33–38
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51(1):69–82
Homeyer N, Gohlke H (2012) Free energy calculations by the molecular mechanics poisson−boltzmann surface area method. Mol Inform 31(2):114–122. https://doi.org/10.1002/minf.201100135
Richmond TJ (1984) Solvent accessible surface area and excluded volume in proteins. Analytical equations for overlapping spheres and implications for the hydrophobic effect. J Mol Biol 178(1):63–89
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10(5):449–461
Acknowledgements
The authors acknowledge the College of Health Science of the University of KwaZulu-Natal for funding and the Centre for High-Performance Computing (CHPC), Cape Town, South Africa, for computational resources (www.chpc.ac.za).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Elamin, G., Aljoundi, A. & Soliman, M.E.S. Multi-catalytic Sites Inhibition of Bcl2 Induces Expanding of Hydrophobic Groove: A New Avenue Towards Waldenström Macroglobulinemia Therapy. Protein J 41, 201–215 (2022). https://doi.org/10.1007/s10930-022-10046-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-022-10046-9