Abstract
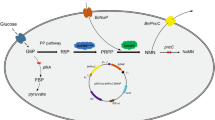
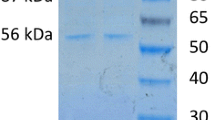
Patients in health-care settings develop nosocomial infections due to prolonged hospital stay. The Gram negative Klebsiella pneumoniae (K. pneumoniae), is a bacterial pathogen responsible for most nosocomial infections and are resistant to most current antibiotics. Hence, there is need for identification and validation of potential protein targets for design of new generation antibiotics. One of such targets is nicotinate nucleotide adenylyltransferase, an enzyme responsible for redox metabolism. This study focuses on novel expression, purification, and biophysical characterization of NNAT from K. pneumoniae. KpNNAT was over-expressed in T7 express™ Escherichia coli using the pGEX-4 T-1 expressions system and purified to > 98% homogeneity (~ 20 mg KpNNAT/g of the wet cell) using a combination of glutathione-agarose and immobilized Ni2+ affinity chromatography. KpNNAT indirectly showed “pseudo-specific activity” of 0.30 μmol/min/mg towards β-nicotinate mononucleotide and ATP using alcohol dehydrogenase as a secondary enzyme (in the presence of ethanol). Far-UV circular dichroism showed a ~ 38% predominantly alpha-helical and 16% β-strand secondary structural content. The binding of ATP to KpNNAT is entropically-driven with an overall ∆G° of ‒23.8 kJ/mol and dissociation constant of 69.1 µM. Data from this study suggest that KpNNAT can be expressed in E. coli, purified to homogeneity to yield high quantities of active recombinant enzyme for downstream biophysical studies such as X-ray crystallography.











Similar content being viewed by others
Data Availability
Not applicable.
Code Availability
Not applicable.
References
Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. The University of Chicago Press; 2008.
Tzouvelekis L, Markogiannakis A, Psichogiou M, Tassios P, Daikos G (2012) Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707
Podschun R, Ullmann U (1998) Klebsiella spp as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 11:589–603
Kaewpoowat Q, Ostrosky-Zeichner L (2015) Tigecycline: a critical safety review. Expert Opin Drug Saf 14:335–342
Ramsamy Y, Essack SY, Sartorius B, Patel M, Mlisana KP (2018) Antibiotic resistance trends of ESKAPE pathogens in Kwazulu-Natal, South Africa: a five-year retrospective analysis. African J Lab Med 7:1–8
Huang N, Kolhatkar R, Eyobo Y, Sorci L, Rodionova I, Osterman AL, MacKerell AD Jr, Zhang H (2010) Complexes of bacterial nicotinate mononucleotide adenylyltransferase with inhibitors: implication for structure-based drug design and improvement. J Med Chem 53:5229–5239
Sorci L, Pan Y, Eyobo Y, Rodionova I, Huang N, Kurnasov O, Zhong S, MacKerell AD Jr, Zhang H, Osterman AL (2009) Targeting NAD biosynthesis in bacterial pathogens: structure-based development of inhibitors of nicotinate mononucleotide adenylyltransferase NadD. Chem Biol 16:849–861
Chaudhary AS, Chen W, Jin J, Tai PC, Wang B (2015) SecA: a potential antimicrobial target. Future Med Chem 7:989–1007
Zawadzke LE, Wu P, Cook L, Fan L, Casperson M, Kishnani M, Calambur D, Hofstead SJ, Padmanabha R (2003) Targeting the MraY and MurG bacterial enzymes for antimicrobial therapeutic intervention. Anal Biochem 314:243–252
Santos JA, Lamers MH (2020) Novel antibiotics targeting bacterial replicative DNA polymerases. Antibiotics 9:776
Guo L, Huang L, Su Y, Qin Y, Zhao L, Yan Q (2018) secA, secD, secF, yajC, and yidC contribute to the adhesion regulation of Vibrio alginolyticus. Microbiologyopen. 7:e00551
Samitha Rao CV, De Waelheyns E, Economou A, Anné J (2014) Antibiotic targeting of the bacterial secretory pathway. Biochimica et Biophysica Acta-Mol Cell Res 1843:1762–83
Xiao W, Wang R-S, Handy DE, Loscalzo J (2018) NAD (H) and NADP (H) redox couples and cellular energy metabolism. Antioxid Redox Signal 28:251–272
Gerdes SY, Scholle MD, D’Souza M, Bernal A, Baev MV, Farrell M, Kurnasov OV, Daugherty MD, Mseeh F, Polanuyer BM (2002) From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol 184:4555–4572
Uddin R, Saeed K (2014) Identification and characterization of potential drug targets by subtractive genome analyses of methicillin resistant Staphylococcus aureus. Comput Biol Chem 48:55–63
Zhang H, Zhou T, Kurnasov O, Cheek S, Grishin NV, Osterman A (2002) Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD. Structure. 10:69–79
Whitmore L, Wallace B (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32:W668–W673
Provencher SW, Gloeckner J (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33–37
Menzella HG (2011) Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli. Microb Cell Fact 10:1–8
Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U, Gileadi O (2008) Codon optimization can improve expression of human genes in Escherichia coli: a multi-gene study. Protein Expr Purif 59:94–102
Kim CH, Oh Y, Lee TH (1997) Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene 199:293–301
Disbrow GL, Sunitha I, Baker CC, Hanover J, Schlegel R (2003) Codon optimization of the HPV-16 E5 gene enhances protein expression. Virology 311:105–114
Bell MR, Engleka MJ, Malik A, Strickler JE (2013) To fuse or not to fuse: what is your purpose? Protein Sci 22:1466–1477
Zhao, X., Li, G., Liang, S.: Several affinity tags commonly used in chromatographic purification. Journal of analytical methods in chemistry. 2013, (2013)
Silverstein TP (2016) The alcohol dehydrogenase kinetics laboratory: enhanced data analysis and student-designed mini-projects. J Chem Educ 93:963–970
Zhai RG, Rizzi M, Garavaglia S (2009) Nicotinamide/nicotinic acid mononucleotide adenylyltransferase, new insights into an ancient enzyme. Cell Mol Life Sci 66:2805–2818
Hawe A, Sutter M, Jiskoot W (2008) Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res 25:1487–1499
Zhang X, Kurnasov OV, Karthikeyan S, Grishin NV, Osterman AL, Zhang H (2003) Structural characterization of a human cytosolic NMN/NaMN adenylyltransferase and implication in human NAD biosynthesis* 210. J Biol Chem 278:13503–13511
Strickland EH, Horwitz J, Billups C (1969) Fine structure in the near-ultraviolet circular dichroism and absorption spectra of tryptophan derivatives and chymotrypsinogen A at 77 K. Biochemistry 8:3205–3213
Strickland EH, Beychok S (1974) Aromatic contributions to circular dichroism spectra of protein. CRC Crit Rev Biochem 2:113–175
Gasymov OK, Abduragimov AR, Glasgow BJ (2014) Probing tertiary structure of proteins using single Trp mutations with circular dichroism at low temperature. J Phys Chem B 118:986–995
Kelly SM, Jess TJ, Price NC (2005) How to study proteins by circular dichroism. Biochimica et Biophysica Acta Proteins Proteomics. 1751:119–39
Mehl RA, Kinsland C, Begley TP (2000) Identification of the Escherichia coli nicotinic acid mononucleotide adenylyltransferase gene. J Bacteriol 182:4372–4374
Yoon H-J, Kim HL, Mikami B, Suh SW (2005) Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation. J Mol Biol 351:258–265
Bathke J, Fritz-Wolf K, Brandstädter C, Burkhardt A, Jortzik E, Rahlfs S, Becker K (2016) Structural and functional characterization of Plasmodium falciparum nicotinic acid mononucleotide adenylyltransferase. J Mol Biol 428:4946–4961
Contreras-Rodríguez LE, Marin-Mogollon CY, Sánchez-Mejía LM, Ramírez-Hernández MH (2018) Structural insights into Plasmodium falciparum nicotinamide mononucleotide adenylyltransferase: oligomeric assembly. Memórias do Instituto Oswaldo Cruz. https://doi.org/10.1590/0074-02760180073
Sershon VC, Santarsiero BD, Mesecar AD (2009) Kinetic and X-ray structural evidence for negative cooperativity in substrate binding to nicotinate mononucleotide adenylyltransferase (NMAT) from Bacillus anthracis. J Mol Biol 385:867–888
Adams PA (1980) The interpretation of entropy/enthalpy compensation phenomena in the deacylation of acyl-α-chymotrypsins. Biochemical Journal 191:653–655
Saridakis V, Christendat D, Kimber MS, Dharamsi A, Edwards AM, Pai EF (2001) Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicumNMN adenylyltransferase complexes. J Biol Chem 276:7225–7232
D’Angelo I, Raffaelli N, Dabusti V, Lorenzi T, Magni G, Rizzi M (2000) Structure of nicotinamide mononucleotide adenylyltransferase: a key enzyme in NAD+ biosynthesis. Structure 8:993–1004
Ladbury JE (1996) Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chem Biol 3:973–980
Acknowledgements
This work was supported by the South African Medical Research Council (SA-MRC) Self-Initiated Research Grant to IA and the South African Research Chairs Initiative (SARChI) of the Department of Science and Technology and National Research Foundation (grant 64788 to IA) and National Research Foundation National Equipment Programme (grant 88047 to Emeritus Prof HW Dirr). TD acknowledges the National Research Foundation (NRF) for funding her MSc degree. RM acknowledges the South African Department of Water and Sanitation for funding her MSc degree. OJ acknowledges the SA-MRC for bursary towards her MSc degree. CA also acknowledges the SARChI programme of the Department of Science and Technology and National Research Foundation for post-doctoral fellowship funding.
Funding
The work was funded by the South African Medical Research Council Self-Initiated Research Grant and the South African Research Chair Initiative of the Department of Science and Technology and National Research Foundation.
Author information
Authors and Affiliations
Contributions
TD, RM and OJ: Investigation, Methodology, Formal analysis, Writing—original draft. CA and TK: Review & editing of manuscript. IA: Writing and Review of manuscript, Supervision, Conceptualisation, Resources and Funding.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest or competing interest.
Ethical Approval
Ethics waiver for this study was awarded to Ikechukwu Achilonu by the Human Research Ethics (Medical) of the University of the Witwatersrand, Ref number: W-CBP-181009-01.
Consent to Participate
Not applicable.
Consent for Publication
All the authors consent to the publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Daya, T., Jeje, O., Maake, R. et al. Expression, Purification, and Biophysical Characterization of Klebsiella Pneumoniae Nicotinate Nucleotide Adenylyltransferase. Protein J 41, 141–156 (2022). https://doi.org/10.1007/s10930-021-10037-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-021-10037-2