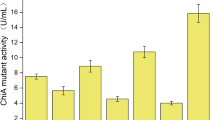
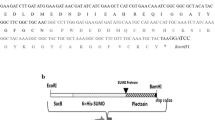
Abstract
Cecropin AD (CAD) is a hybrid peptide composed of 37 amino acids with the characters of strong antibacterial, antitumor properties and no hemolytic activity, which was regarded as a promising antibiotic candidate. Thus, a safe method to produce Cecropin AD is necessary to be found. In the study, Bacillus subtilis WB800N was employed as host strain. The CAD coding sequence fused with the signal peptide of SPsacB, the 6 × His gene and the gene of small ubiquitin-like modifier were cloned into the maltose-inducible vector pGJ148. Under the induction by 6% maltose, the recombinant fusion protein was successfully expressed and detected in culture substrate. An optimized amount (26.4 mg/L) of the recombinant CAD was purified of culture supernatant. After purification and digestion, the recombinant CAD was harvested about 4.5 mg/L with a purity of 93%. Recombinant CAD exhibited similar antimicrobial activity with synthetic CAD. This shows that the production of CAD in maltose-induced Bacillus subtilis expression system is a relatively safe method, which is vital for the application of CAD in animal husbandry production.






Similar content being viewed by others
Data Availability
All data generated or analysed in the study are included in the published article.
References
Van Boeckel TP, Brower C, Gilbert M et al (2015) Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA 112:5649–5654. https://doi.org/10.2307/26462640
Oloso N, Fagbo S, Garbati M, et al (2018) Antimicrobial resistance in food animals and the environment in Nigeria: A Review. Inter J Env Res Pub Heal 15 (6): 1284. https://doi.org/10.3390/ijerph15061284
Wu Q, Patočka J, Kuča K (2018) Insect antimicrobial peptides, a mini review. Toxins 10(11):461. https://doi.org/10.3390/toxins10110461
Ahmed TAE, Hammami R (2018) Recent insights into structure-function relationships of antimicrobial peptides. J Food Biochem. https://doi.org/10.1111/jfbc.12546
Yang K, Su Y, Li J, Sun J, Yang Y (2012) Expression and purification of the antimicrobial peptide cecropin AD by fusion with cationic elastin-like polypeptides. Protein Expres Purif 85(2):200–203. https://doi.org/10.1016/j.pep.2012.04.007
Zhu W, Gong G, Pan J, Han S, Zhang W, Hu Y, Xie L (2018) High level expression and purification of recombinant human serum albumin in Pichia pastoris. Protein Expres Purif 147:61–68. https://doi.org/10.1016/j.pep.2018.02.003
Wibowo D, Zhao CX (2018) Recent achievements and perspectives for large-scale recombinant production of antimicrobial peptides. Appl Microbiol Biot. https://doi.org/10.1007/s00253-018-9524-1
Ovaa H, Vertegaal ACO (2018) Probing ubiquitin and SUMO conjugation and deconjugation. Biochem Soc T 46(2):423–436. https://doi.org/10.1042/bst20170086
Praefcke GJK, Hofmann K, Dohmen RJ (2012) SUMO playing tag with ubiquitin. Trends Biochem Sci 37(1):23–31. https://doi.org/10.1016/j.tibs.2011.09.002
Zhang J, Sun A, Dong Y, Wei D (2017) Recombinant production and characterization of SAC, the core domain of Par-4, by SUMO fusion system. Appl Biochem Biotech 184 (4): 1155-1167. https://doi.org/10.1007/s12010-017-2599-9
Kota V, Sommer G, Hazard ES, Hardiman G, Twiss JL, Heise T (2017) SUMO modification of the RNA-binding protein la regulates cell proliferation and STAT3 protein stability. Mol Cell Biol. https://doi.org/10.1128/mcb.00129-17
Namvar S, Barkhordari F, Raigani M, Jahandar H, Nematollahi L, Davami F (2018) Cloning and soluble expression of mature α-luffin from Luffa cylindrica in E. coli using SUMO fusion protein. Turk J Biol 42:23–32. https://doi.org/10.3906/biy-1708-12
Mukhopadhyay D, Dasso M (2007) Modification in reverse: the SUMO proteases. Trends Biochem Sci 32(6):286–295. https://doi.org/10.1016/j.tibs.2007.05.002
Shimokawa-Falcão L, Caporrino M, Barbaro K, Della-Casa M, Magalhães G (2017) Toxin Fused with SUMO Tag: A new expression vector strategy to obtain recombinant venom toxins with easy tag removal inside the bacteria. TOXINS 9 (3): 82. https://doi.org/10.3390/toxins9030082
Lefevre M, Racedo SM, Denayrolles M et al (2017) Safety assessment of Bacillus subtilis CU1 for use as a probiotic in humans. Regul Toxicol Pharm 83:54–65. https://doi.org/10.1016/j.yrtph.2016.11.010
Zuenko VA, Laktionov KS, Pravdin IV, Kravtsova LZ, Ushakova NA (2017) Effect of Bacillus subtilis in feed probiotic on the digestion of fish cultured in cages. J Ichthyol 57(1):152–157. https://doi.org/10.1134/s0032945217010143
Hmani H, Daoud L, Jlidi M, et al (2017) A Bacillus subtilis strain as probiotic in poultry: selection based on in vitro functional properties and enzymatic potentialities. J Ind Microbiol Biot 44 (8): 1157-1166. https://doi.org/10.1007/s10295-017-1944-x
Song Y, Nikoloff JM, Fu G, et al (2016) Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications. PLOS ONE 11 (7): e0158447. https://doi.org/10.1371/journal.pone.0158447
Zhou C, Ye B, Cheng S, Zhao L, Liu Y, Jiang J, Yan X (2019) Promoter engineering enables overproduction of foreign proteins from a single copy expression cassette in Bacillus subtilis. Microb Cell Fact. https://doi.org/10.1186/s12934-019-1159-0
Gu Y, Xu X, Wu Y, et al (2018) Advances and prospects of Bacillus subtilis cellular factories: From rational design to industrial applications. Metab Eng. https://doi.org/10.1016/j.ymben.2018.05.006
Barns KJ, Weisshaar JC (2016) Single-cell, time-resolved study of the effects of the antimicrobial peptide alamethicin on Bacillus subtilis. BBA-Biomembranes 1858 (4): 725-732. https://doi.org/10.1016/j.bbamem.2016.01.003
Ji S, Li W, Baloch AR, Wang M, Li H, Cao B, Zhang H (2017) Efficient biosynthesis of a cecropin A-melittin mutant in Bacillus subtilis WB700. Sci Rep UK. https://doi.org/10.1038/srep40587
Phan TTP, Nguyen HD, Schumann W (2006) Novel plasmid-based expression vectors for intra- and extracellular production of recombinant proteins in Bacillus subtilis. Protein Expres Purif 46(2):189–195. https://doi.org/10.1016/j.pep.2005.07.005
Phan TTP, Nguyen HD, Schumann W (2012) Development of a strong intracellular expression system for Bacillus subtilis by optimizing promoter elements. J Biotechnol 157(1):167–172. https://doi.org/10.1016/j.jbiotec.2011.10.006
Morabbi Heravi K, Wenzel M, Altenbuchner J (2011) Regulation of mtl operon promoter of Bacillus subtilis: requirements of its use in expression vectors. Microb Cell Fact 10(1):83. https://doi.org/10.1186/1475-2859-10-83
Zhang XZ, Cui ZL, Hong Q, Li SP (2005) High-Level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl Environ Microb 71 (7): 4101-4103. https://doi.org/10.1128/aem.71.7.4101-4103.2005
Ming YM, Wei ZW, Lin CY, Sheng GY (2010) Development of a Bacillus subtilis expression system using the improved Pglv promoter. Microb Cell Fact 9(1):55. https://doi.org/10.1186/1475-2859-9-55
Sun H, Bie X, Lu F, Lu Y, Wu Y, Lu Z (2009) Enhancement of surfactin production of Bacillus subtilis fmbR by replacement of the native promoter with the Pspac promoter. Can J Microbiol 55(8):1003–1006. https://doi.org/10.1139/w09-044
Liu H, Liu H, Yang S, Wang R, Wang T (2019) Improved expression and optimization of trehalose synthase by regulation of Pglv in Bacillus subtilis. Sci Rep. https://doi.org/10.1038/s41598-019-43172-z
Xue GP, Johnson JS, Dalrymple BP (1999) High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J Microbiol Meth 34 (3): 183-191
Dong N, Zhu X, Chou S, Shan A, Li W, Jiang J (2014) Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials 35:8028–8039
Stark M, Liu LP, Deber CM (2002) Cationic hydrophobic peptides with antimicrobial activity. Antimicrob Agents Ch 46:3585–3590. https://doi.org/10.1128/AAC.46.11.3585-3590.2002
Wu S, Zhang F, Huang Z et al (2012) Effects of the antimicrobial peptide cecropin AD on performance and intestinal health in weaned piglets challenged with Escherichia coli. Peptides 35(2):225–230. https://doi.org/10.1016/j.peptides.2012.03.030
Cui X, Jiang Y, Chang L, Meng L, Yu J, Wang C, Jiang X (2018) Heterologous expression of an agarase gene in Bacillus subtilis, and characterization of the agarase. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2018.07.118
Zhang L, Li G, Zhan N, Sun T, Cheng B, Li Y, Shan A (2019) Expression of a Pseudomonas aeruginosa-targeted antimicrobial peptide T9W in Bacillus subtilis using a maltose-inducible vector. Process Biochem. https://doi.org/10.1016/j.procbio.2019.03.008
Fu LH, Wang Y, Ju JS, et al (2019) Extracellular production of active-form Streptomyces mobaraensis transglutaminase in Bacillus subtilis. Appl Microbiol Biot. https://doi.org/10.1007/s00253-019-10256-9
Zhang LC, Wei DD, Zhan N et al (2020) Heterologous expression of the novel α-helical hybrid peptide PR-FO in Bacillus subtilis. Bioproc Biosyst Eng. https://doi.org/10.1007/s00449-020-02353-1
Lin HJ, Xiao Joe JT, Lu WJ, et al (2020) Secretory Production of Functional Grouper Type I Interferon from Epinephelus septemfasciatus in Escherichia coli and Bacillus subtilis. Int J Mol Sci 21 (4) : 1465. https://doi.org/10.3390/ijms21041465
Wang Y, Liu Y, Wang Z et al (2014) Influence of promoter and signal peptide on the expression of pullulanase in Bacillus subtilis. Biotechnol Lett 238:41926–41936
Li W, Zhou X, Lu P (2004) Bottlenecks in the expression and secretion of heterologous proteins in Bacillus subtilis. Res Microbiol 155(8):605–610
Petersen TN, Brunak S, von Heijne G et al (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786
Jin FL, Xu XX, Yu XQ, Ren SX (2009) Expression and characterization of antimicrobial peptide Cecropin AD in the methylotrophic yeast. Pichia Pastoris Process Biochem 44:11–16
Chen X, Zhu FM, Cao YH, Qiao SY (2009) Novel expression vector for secretion of cecropin AD in Bacillus subtilis with enhanced antimicrobial activity. Antimicrob Agents Chem 53: 3683–3689
Funding
This work was supported by the Grants form Natural Science Foundation of China (31672434, 31872368, 31472104), the China Agriculture Research System (CARS-35), and the Natural Science Foundation of Heilongjiang Province (TD2019C001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared that they had no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, L., Li, X., Zhan, N. et al. Maltose Induced Expression of Cecropin AD by SUMO Technology in Bacillus subtilis WB800N. Protein J 39, 383–391 (2020). https://doi.org/10.1007/s10930-020-09908-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09908-x