Abstract
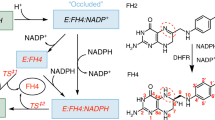
The enzyme dihydrofolate reductase (DHFR) catalyzes NADPH dependent reduction of dihydrofolate to tetrahydrofolate. It plays a crucial role in the DNA synthesis. The investigation of evolution of DHFR generates immense curiosity. It aids in predicting how the enzyme has adapted to the surroundings of various cell types. In spite of great similarity in the structure of E. coli DHFR and human DHFR, their primary sequences are divergent to a great extent, which is evident in variations in the kinetics mechanism of their catalysis. In presence of physiological levels of ligands, they possess distinct kinetics and different rate limiting steps. We have reviewed the process of their unfolding and refolding, their behaviour in denaturing conditions and in presence of various chaperones. Although there is structural similarity between these two homologous enzymes yet they have established distinct mechanisms to accomplish the coequal functions.




Similar content being viewed by others
References
Chen YQ, Kraut J, Callender R (1997) pH-dependent conformational changes in Escherichia coli dihydrofolate reductase revealed by raman difference spectroscopy. Biophys J 72:936–941
Kuwajima K, Garvey EP, Finn BE, Matthews CR, Sugai S (1991) Transient intermediates in the folding of dihydrofolate reductase as detected by far-ultraviolet circular dichroism spectroscopy. Biochemistry 30(31):7693–7703
Liu CT, Francis K, Layfield JP, Huang X, Schiffer SH, Kohen A, Benkovic SJ (2014) Escherichia coli dihydrofolate reductase catalyzed proton and hydride transfers: temporal order and the roles of Asp27 and Tyr100. PNAS 111:18231–18236
Rashid N, Thapliyal C, Chaudhuri P (2018) Quantification of differential efficacy of chemical chaperones in ameliorating solubilization and folding of zebrafish dihydrofolate reductase. Int J of Biol Macromol 111:186–192
Blakley RL (1995) Eukaryotic dihydrofolate reductase. Adv Enzymol Relat Areas Mol Biol 70:23–102
Rashid N., Thapliyal C., Chaudhuri P. (2016). Dihydrofolate reductase as a versatile drug target in health care. J Proteins Proteomics 7(4), 247–257.
Wallace LA, Matthews CR (2002) Highly divergent dihydrofolate reductases conserve complex folding mechanisms. J Mol Biol 315:193–211
Davies JF 2nd, Delcamp TJ, Prendergast NJ, Ashford VA, Freisheim JH, Kraut J (1990) Crystal structures of recombinant human dihydrofolate reductase complexed with folate and 5-deazafolate. Biochemistry 29:9467–9479
Sawaya MR, Kraut J (1997) Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry 36:586–603
Bhabha G., Ekiert D.C., Jennewein M., Zmasek C.M., Tuttle L.M., Kroon G., Dyson J., Godzik A., Wilson I.A., Wright P.E. (2013). Divergent evolution of protein conformational dynamics in dihydrofolate reductase Nat Struct Mol Biol 20:1243–1249.
Benkovic SJ, Hammes-Schiffer S (2003) A perspective on enzyme catalysis. Science 301:1196–1202
Schnell JR, Dyson HJ, Wright PE (2004) Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct 33:119–140
Hammes-Schiffer S, Benkovic SJ (2006) Relating protein motion to catalysis. Annu Rev Biochem 75:519–541
Thapliyal C, Jain N, Chaudhuri (Chattopadhyay) P (2016) Investigation of folding unfolding process of a new variant of dihydrofolate reductase protein from Zebrafish. Int J Biol Macromol 91:736–743
Shrimpton P, Mullaney A, Allemann RK (2003) Functional role for Tyr 31 in the catalytic cycle of chicken dihydrofolate reductase. Proteins 51:216–223
Kasper JR, Liu PF, Park C (2014) Structure of a partially unfolded form of Escherichia coli dihydrofolate reductase provides insight into it’s folding pathway. Protein Sci 23(12):1728–1737
Sham YY, Ma B, Tsai CJ, Nussinov R (2001) Molecular dynamics simulation of Escherichia coli dihydrofolate reductase and its protein fragments: relative stabilities in experiment and simulations. Protein Sci 10(1):135–148
Pan H, Lee JC, Hilser VJ (2000) Binding sites in Escherichia coli dihydrofolate reductase communicate by modulating the conformational ensemble. PNAS 97(22):12020–12025
Mittal J, Ravitchandirane G, Chaudhuri TK, Chaudhuri P (2010) Structure-activity correlationship and folding of recombinant Escherichia coli dihydro folate reductase (DHFR) enzyme through biochemical and biophysical approaches. J Biophys Chem 1(2):105–112
Martin J, Langer T, Boteva R, Schramel A, Horwich AL, Hartl FU (1991) Chaperonin-mediated protein folding at the surface of GroEL through a ‘molten globule’-like intermediate. Nature 352(6330):36–42
Viitanen PV, Gatenby AA, Lorimer GH (1992) Purified chaperonin 60 (GroEL) interacts with the nonnative states of a multitude of Escherichia coli proteins. Protein Sci 1:363–369
Mayhew M, da Silva AC, Martin J, Erdjument-Bromage H, Tempst P, Hartl FU (1996) Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature 379(6564):420–426
Horst R, Fenton WA, EnglandeR SW, Wuthrich K, Horwich AL (2007) Folding trajectories of human dihydrofolate reductase inside the GroEL–GroES chaperonin cavity and free in solution. PNAS 104(52):20788–20792
Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wuthrich K (2005) Direct NMR observation of a substrate protein bound to the chaperonin GroEL. PNAS 102(36):12748–12753
Goldberg MS, Zhang J, Sondek S, Mathews CR, Fox RO, Horwich AL (1997) Native-like structure of a protein-folding intermediate bound to the chaperonin GroEL. Biochemistry 94:1080–1085
Rashid N, Thapliyal C, Chaudhuri P (2017) Osmolyte induced enhancement of expression and solubility of human dihydrofolate reductase: an in vivo study. Int J Biol Macromol 103:1044–1053
Frieden C (1990) Refolding of Escherichia coli dihydrofolate reductase: sequential formation of substrate binding sites. PNAS 87(12):4413–4416
Thapliyal C, Jain N, Chaudhuri P (2015) Comparison of physico-chemical aspects between E. coli and human dihydrofolate reductase: an equilibrium unfolding study. Biophysics 60:378–386
Baker DJ, Beddell CR, Champness JN, Goodford PJ, Norrington FEA, Smith DR, Stammers DK (1981) The binding of trimethoprim to bacterial dihydrofolate reductase. FEBS Lett 126:49–52
Acknowledgements
The authors acknowledge the financial assistance provided by Amity Institute of Biotechnology, Amity University, Noida, Uttar Pradesh, India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rashid, N., Chaudhuri (Chattopadhyay), P. Evolutionarily Related Dihydrofolate Reductases Perform Coequal Functions Yet Show Divergence in Their Trajectories. Protein J 37, 301–310 (2018). https://doi.org/10.1007/s10930-018-9784-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-018-9784-8