Abstract
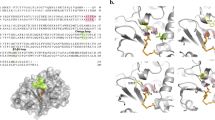
Escherichia coli PBP5, a DD-carboxypeptidase (DD-CPase), helps in maintaining cell shape and intrinsic β-lactam resistance. Though PBP5 does not have β-lactamase activity under physiological pH, it has a common but shorter Ω-like loop resembling class A β-lactamases. However, such Ω-like loop lacks the key glutamic acid residue that is present in β-lactamases. It is speculated that β-lactamases and DD-CPases might have undergone divergent evolution leading to distinct enzymes with different substrate specificities and functions indicating the versatility of the Ω-loops. Nonetheless, direct experimental evidence favoring the idea is insufficient. Here, aiming to investigate the effect of introducing a glutamic acid residue in the PBP5 Ω-like loop, we substituted A184 to E to create PBP5_A184E. Expression of PBP5_A184E in E. coli ∆PBP5 mutant elevates the β-lactam resistance, especially for cephalosporins. However, like PBP5, PBP5_A184E has the ability to complement the aberrantly shaped E. coli septuple PBP mutant indicating an unaffected in vivo DD-CPase activity. Biochemical and bioinformatics analyses have substantiated the dual enzyme nature of the mutated enzyme possessing both DD-CPase and β-lactamase activities. Therefore, substitution of A184 to E of Ω-like loop alone can introduce the cephalosporinase activity in E. coli PBP5 supporting the phenomenon of a single amino acid polymorphism.




Similar content being viewed by others
References
Ghuysen J-M (1991) Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol 45(1):37–67
Ghosh AS, Chowdhury C, Nelson DE (2008) Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol 16(7):309–317
Gonzalez-Leiza SM, de Pedro MA, Ayala JA (2011) AmpH, a bifunctional DD-endopeptidase and DD-carboxypeptidase of Escherichia coli. J Bacteriol 193(24):6887–6894. https://doi.org/10.1128/JB.05764-11
Sarkar SK, Chowdhury C, Ghosh AS (2010) Deletion of penicillin-binding protein 5 (PBP5) sensitises Escherichia coli cells to β-lactam agents. Int J Antimicrob Agents 35(3):244–249
Nelson DE, Young KD (2000) Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol 182(6):1714–1721
Peters K, Kannan S, Rao VA, Biboy J, Vollmer D, Erickson SW, Lewis RJ, Young KD, Vollmer W (2016) The redundancy of peptidoglycan carboxypeptidases ensures robust cell shape maintenance in Escherichia coli. MBio 7(3):e00819
Nelson DE, Young KD (2001) Contributions of PBP 5 anddd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J Bacteriol 183(10):3055–3064
Sarkar SK, Dutta M, Chowdhury C, Kumar A, Ghosh AS (2011) PBP5, PBP6 and DacD play different roles in intrinsic β-lactam resistance of Escherichia coli. Microbiology 157(9):2702–2707
Nicholas RA, Strominger JL (1988) Site-directed mutants of a soluble form of penicillin-binding protein 5 from Escherichia coli and their catalytic properties. J Biol Chem 263(4):2034–2040
Urbach C, Fastrez J, Soumillion P (2008) A new family of cyanobacterial penicillin-binding proteins a missing link in the evolution of class a β-lactamases. J Biol Chem 283(47):32516–32526
Dutta M, Kar D, Bansal A, Chakraborty S, Ghosh AS (2015) A single amino acid substitution in the Ω-like loop of E. coli PBP5 disrupts its ability to maintain cell shape and intrinsic beta-lactam resistance. Microbiology 161(4):895–902
Gerlt JA, Babbitt PC (2001) Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem 70(1):209–246
Nicholas RA, Krings S, Tomberg J, Nicola G, Davies C (2003) Crystal structure of wild-type penicillin-binding protein 5 from Escherichia coli implications for deacylation of the acyl-enzyme complex. J Biol Chem 278(52):52826–52833
Chowdhury C, Nayak TR, Young KD, Ghosh AS (2010) A weak DD-carboxypeptidase activity explains the inability of PBP 6 to substitute for PBP 5 in maintaining normal cell shape in Escherichia coli. FEMS Microbiol Lett 303(1):76–83
Chambers HF, Sachdeva MJ, Hackbarth CJ (1994) Kinetics of penicillin binding to penicillin-binding proteins of Staphylococcus aureus. Biochem J 301(1):139–144
Di Guilmi AM, Mouz N, Pétillot Y, Forest E, Dideberg O, Vernet T (2000) Deacylation kinetics analysis of Streptococcus pneumoniae penicillin-binding protein 2x mutants resistant to β-lactam antibiotics using electrospray ionization–mass spectrometry. Anal Biochem 284(2):240–246
Frére J-M, Leyh-Bouille M, Ghuysen J-M, Nieto M, Perkins HR (1976) [51] Exocellular dd-carboxypeptidases-transpeptidases from Streptomyces. Methods Enzymol 45:610–636
Chowdhury C, Kar D, Dutta M, Kumar A, Ghosh AS (2012) Moderate deacylation efficiency of DacD explains its ability to partially restore beta-lactam resistance in Escherichia coli PBP5 mutant. FEMS Microbiol Lett 337(1):73–80
Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C (2012) Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 80(2):374–381
Rost B, Yachdav G, Liu J (2004) The predictprotein server. Nucleic Acids Res 32(suppl 2):W321–W326
Nilsen T, Ghosh AS, Goldberg MB, Young KD (2004) Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol Microbiol 52(4):1045–1054
Banerjee S, Pieper U, Kapadia G, Pannell LK, Herzberg O (1998) Role of the Ω-Loop in the activity, substrate specificity, and structure of class a β-lactamase. Biochemistry 37(10):3286–3296
Sun T, Nukaga M, Mayama K, Braswell EH, Knox JR (2003) Comparison of β-lactamases of classes a and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci 12(1):82–91
Ropy A, Cabot G, Sánchez-Diener I, Aguilera C, Moya B, Ayala JA, Oliver A (2015) Role of Pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, β-Lactam resistance, and peptidoglycan structure. Antimicrob Agents Chemother 59(7):3925–3934
Smith JD, Kumarasiri M, Zhang W, Hesek D, Lee M, Toth M, Vakulenko S, Fisher JF, Mobashery S, Chen Y (2013) Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in β-lactam resistance. Antimicrob Agents Chemother 57(7):3137–3146
Haenni M, Moreillon P (2006) Mutations in penicillin-binding protein (PBP) genes and in non-PBP genes during selection of penicillin-resistant Streptococcus gordonii. Antimicrob Agents Chemother 50(12):4053–4061
Terao Y, Miyamoto K, Ohta H (2006) Introduction of single mutation changes arylmalonate decarboxylase to racemase. Chem Commun 34:3600–3602
Seebeck FP, Guainazzi A, Amoreira C, Baldridge KK, Hilvert D (2006) Stereoselectivity and expanded substrate scope of an engineered PLP-dependent aldolase. Angew Chem Int Ed 45(41):6824–6826
Seebeck FP, Hilvert D (2003) Conversion of a PLP-dependent racemase into an aldolase by a single active site mutation. J Am Chem Soc 125(34):10158–10159
Davies C, White SW, Nicholas RA (2001) Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3-Å resolution. J Biol Chem 276(1):616–623
Stefanova ME, Davies C, Nicholas RA, Gutheil WG (2002) pH, inhibitor, and substrate specificity studies on Escherichia coli penicillin-binding protein 5. Biochim et Biophys Acta 1597(2):292–300
Chowdhury C, Ghosh AS (2011) Differences in active-site microarchitecture explain the dissimilar behaviors of PBP5 and 6 in Escherichia coli. J Mol Graph Model 29(5):650–656
Zhang W, Shi Q, Meroueh SO, Vakulenko SB, Mobashery S (2007) Catalytic mechanism of penicillin-binding protein 5 of Escherichia coli. Biochemistry 46(35):10113–10121
Malhotra KT, Nicholas RA (1992) Substitution of lysine 213 with arginine in penicillin-binding protein 5 of Escherichia coli abolishes d-alanine carboxypeptidase activity without affecting penicillin binding. J Biol Chem 267(16):11386–11391
Shi Q, Meroueh SO, Fisher JF, Mobashery S (2008) Investigation of the mechanism of the cell wall DD-carboxypeptidase reaction of penicillin-binding protein 5 of Escherichia coli by quantum mechanics/molecular mechanics calculations. J Am Chem Soc 130(29):9293–9303
Adachi H, Ohta T, Matsuzawa H (1991) Site-directed mutants, at position 166, of RTEM-1 beta-lactamase that form a stable acyl-enzyme intermediate with penicillin. J Biol Chem 266(5):3186–3191
Escobar WA, Tan AK, Fink AL (1991) Site-directed mutagenesis of. beta.-lactamase leading to accumulation of a catalytic intermediate. Biochemistry 30(44):10783–10787
Strynadka NC, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MN (1992) Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature 359(6397):700–705
Nemmara VV, Dzhekieva L, Subarno Sarkar K, Adediran S, Duez C, Nicholas RA, Pratt RF (2011) Substrate specificity of low-molecular mass bacterial DD-peptidases. Biochemistry 50(46):10091–10101
Stec B, Holtz KM, Wojciechowski CL, Kantrowitz ER (2005) Structure of the wild-type TEM-1 β-lactamase at 1.55 Å and the mutant enzyme Ser70Ala at 2.1 Å suggest the mode of noncovalent catalysis for the mutant enzyme. Acta Crystallogr Sect D 61(8):1072–1079
Leszczynski JF, Rose GD (1986) Loops in globular proteins: a novel category of secondary structure. Science 234(4778):849–855
Peimbert M, Segovia L (2003) Evolutionary engineering of a β-lactamase activity on a D-Ala D-Ala transpeptidase fold. Protein Eng 16(1):27–35
Gordon E, Mouz N, Duee E, Dideberg O (2000) The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J Mol Biol 299(2):477–485
Pares S, Mouz N, Petillot Y, Hakenbeck R, Dideberg O (1996) X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat Struct Mol Biol 3(3):284–289
Bansal A, Kar D, Murugan RA, Mallick S, Dutta M, Pandey SD, Chowdhury C, Ghosh AS (2015) A putative low-molecular-mass penicillin-binding protein (PBP) of Mycobacterium smegmatis exhibits prominent physiological characteristics of DD-carboxypeptidase and beta-lactamase. Microbiology 161(5):1081–1091
Acknowledgements
This work is supported by a grant from Department of Biotechnology, Govt. of India (grant # BT/PR8539/BRB/10/1235/2013) to ASG. DK and MD are supported by Indian Institute of Technology Kharagpur. SDP thanks, Science and Engineering Research Board (SERB), Govt. of India for National Post-doctoral Fellowship vide grant # PDF/2015/000011.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that there exists no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kar, D., Pandey, S.D., Mallick, S. et al. Substitution of Alanine at Position 184 with Glutamic Acid in Escherichia coli PBP5 Ω-Like Loop Introduces a Moderate Cephalosporinase Activity. Protein J 37, 122–131 (2018). https://doi.org/10.1007/s10930-018-9765-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-018-9765-y