Abstract
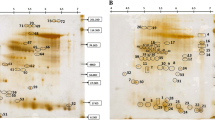
Low temperature can greatly restrict the growth and development of rice. The rice seedlings show growth retardation, lamina wrap, and part of blade even died under the condition of low temperature. In order to get more information about cold stress responses in rice, two dimensional electrophoresis and bioinformatics analysis of mass spectrometry were used to preliminary survey the cold tolerance of cold sensitive line 9311 and cold resistance variety Fujisaka 5 under cold stress. Two dimensional electrophoresis maps of 9311 and Fujisaka 5 were established under cold treatment. With analysis of bioinformation, the proteins were found involve in many aspects of rice development. The largest category of proteins is functioning on metabolism. By comparing the proteins from the two varieties, it can be found that most proteins from 9311 were down-regulated and were up-regulated in Fujisaka 5. The results showed that the membrane composition and structure were damaged, metabolism changed dramatically and rice defense system was activated under the cold stimulation. Fifty-nine proteins related to the resistance of cold stress were identified in our study, and we have investigated and classified all of their biological functions. The importance of our study are providing some conduct for the research of rice resistant to cold stress, supporting auxiliary technique for rice varieties and widening the search field of cold tolerance in plants.



Similar content being viewed by others
Abbreviations
- 2-DE:
-
Two dimensional electrophoresis
- MS:
-
Mass spectrometry
- iTRAQ:
-
Isobaric tags for relative and absolute quantitation
- Rubisco LSU:
-
Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit
- IPG:
-
Immobilized pI gradient
- IEF:
-
Isoelectric focusing
- MALDI TOF:
-
Matrix-assisted laser desorption ionization time-of-flight
References
Cui SX, Huang F, Wang J, Ma X, Cheng Y, Liu J (2005) A proteomic analysis of cold stress responses in rice seedlings. Proteomics 5:3162–3172
Imin N, Kerim T, Rolfe BG, Weinman J (2004) Effect of early cold stress on the maturation of rice anthers. Proteomics 4:1873–1882
Hayashi T, Yamaguchi T, Nakayama K, Komatsu S, Koike S (2006) Susceptibility to coolness at the young microspore stage under high nitrogen supply in rice (Oryza sativa L.). Proteome analysis of mature anthers. Plant Prod Sci 9:212–218
Vergnolle C, Vaultier MN, Taconnat L, Renou JP, Kader JC, Zachowski A, Ruelland E (2005) The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol 139:1217–1233
Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17:3155–3175
Buchanan CD, Lim S, Salzman RA, Kagiampakis I, Morishige DT, Weers BD, Klein RR, Pratt LH, Cordonnier-Pratt MM, Klein PE, Mullet JE (2005) Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Mol Biol 58:699–720
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, ItoY, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stress and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Kawamura Y, Uemura M (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36:141–154
Xu J, Li Y, Sun J, Du L, Zhang Y, Yu Q, Liu X (2013) Comparative physiological and proteomic response to abrupt low temperature stress between two winter wheat cultivars differing in low temperature tolerance. Plant Biol 15:292–303
Yang QS, Wu JH, Li CY, Wei YR, Sheng O, Hu CH, Kuang RB (2012) Quantitative proteomic analysis reveals that antioxidation mechanisms contribute to cold tolerance in plantain (Musa paradisiaca L.; ABB Group) seedlings. Mol Cell Proteom 11:1853–1869
Komatsu S, Karibe H, Hamada T, Rakwal R (1999) Phosphorylation upon cold stress in rice (Oryza sativa L.) seedlings. Theor Appl Genet 9:1304–1310
Tsugita A, Kamo M (1999) 2-D electrophoresis of plant proteins. Methods Mol Biol 112:95–97
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Wessel D, Flugge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143
Huang F, Parmryd I, Nilsson F, Persson AL, Pakrasi HB, Andersson B, Norling B (2002) Proteomics of Synechocystis sp. strain PCC 6803 identification of plasma membrane proteins. Mol Cell Proteom 1:956–966
Fulda S, Huang F, Nilsson F, Hagemann M, Norling B (2000) Proteomics of Synechocystis sp. strain PCC 6803 identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur J Biochem 267:5900–5907
Jin BF, He K, Wang HX, Wang J, Zhou T, Lan Y, Hu MR, Wei KH, Yang SC, Shen BF, Zhang XM (2003) Proteomic analysis of ubiquitin-proteasome effects: insight into the function of eukaryotic initiation factor 5 A. Oncogene 22:4819–4830
Jie Zou, Cuifang Liu, Xinbo Chen (2011) Proteomics of rice in response to heat stress and advances in genetic engineering for heat tolerance in rice. Plant Cell Rep 30:2155–2165
Gammulla CG, Pascovici D, Atwell BJ, Haynes PA (2011) Differential proteomic response of rice (Oryza sativa) leaves exposed to high- and low-temperature stress. Proteomics 11:2839–2850
Yang PF, Li XJ, Liang Y, Jing YX, Shen SH, Kuang TY (2006) Proteomic analysis of the response of Liangyoupeijiu (super high-yield hybrid rice) seedlings to cold stress. J Integr Plant Biol 48(8):945–951
Ma Y, Dai XY, Xu YY, Luo W, Zheng XM, Zeng D, Pan YJ, Lin XL, Liu HH, Zhang DJ, Xiao J, Guo XY, Xu SJ, Niu YD, Jin JB, Zhang H, Xu X, Li LG, Wang W, Qian Q, Ge S, Chong K (2015) COLD1 confers chilling tolerance in rice. Cell 160(6):1209–1221
Chen XH, Zhang WF, Zhang BQ, Zhou JC, Wang YF, Yang QB, Ke YQ, He HQ (2011) Phosphoproteins regulated by heat stress in rice leaves. Proteom Sci 9:37–45
Makoto Hashimoto, Setsuko Komatsu (2007) Proteomic analysis of rice seedlings during cold stress. Proteomics 7:1293–1302
Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteom 5:484–496
Lee DG, Ahsan N, Lee JJ, Bahk JD, Kang KY, Lee BH (2009) Chilling stress-induced proteomic changes in rice roots. J Plant Physiol 166:1–11
Neilson KA, Mariani M, Haynes PA (2011) Quantitative proteomic analysis of cold-responsive proteins in rice. Proteomics 11:1696–1706
Zhang ZG, Zhang QA, Wu JX, Zheng X, Zheng S, Sun XH, Qiu QS, Lu TG (2013) Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 8(2):e57472
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Chen JH, Tian L, Xu HF, Tian DG, Luo YM, Ren CM, Yang LM, Shi JS (2012) Cold-induced changes of protein and phosphoprotein expression patterns from rice roots as revealed by multiplex proteomic analysis. Plant Omics J 5(2):194–199
Acknowledgements
This work was supported by the Outstanding Young Teacher Training Project in High Education Institutions of Guangxi Province (No. GXQG022014003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human Participants and Animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Ji, L., Zhou, P., Zhu, Y. et al. Proteomic Analysis of Rice Seedlings Under Cold Stress. Protein J 36, 299–307 (2017). https://doi.org/10.1007/s10930-017-9721-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-017-9721-2