Abstract
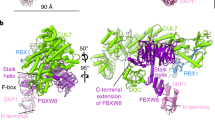
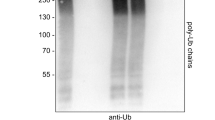
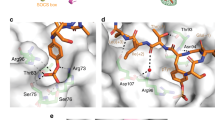
Human ankyrin repeat and suppressor of cytokine signaling box protein 9 (hASB9) is a specific substrate-recognition subunit of an elongin C-cullin-SOCS box E3 ubiquitin ligase complex. It recognizes its substrate, brain type creatine kinase (CKB), using the ankyrin repeat domain; and facilitates the polyubiquitination of CKB to mediate proteasomal degradation through the SOCS box domain. HASB9-2 is an isoform of hASB9 that contains one ankyrin repeat domain. In this study, the crystal structure of hASB9-2 is shown at 2.2-Å resolution using molecular replacement. Overall, hASB9-2 forms a slightly curved arch with a characteristic L-shaped cross-section. Amino acid substitution analysis based on docking experiments revealed that His103 and Phe107 in hASB9-2 are essential for binding to CKB. Analysis of truncation mutants demonstrated that the first six ankyrin repeats along with the N-terminal region of hASB9-2 contribute to the interaction with CKB.





Similar content being viewed by others
Abbreviations
- hASB9:
-
Human ankyrin repeat and suppressor of cytokine signaling box protein 9
- ASBs:
-
Ankyrin repeat and suppressor of cytokine signaling box proteins
- SOCS:
-
Suppressor of cytokine signaling
- ECS:
-
Elongin C-cullin-SOCS box
- CKB:
-
Brain type creatine kinase
- CK:
-
Creatine kinase
- E1:
-
Ubiquitin-activating enzyme
- E2:
-
Ubiquitin-conjugating enzyme
- PDB:
-
Protein Data Bank
- GST:
-
Glutathione S-transferase
- PCR:
-
Polymerase chain reaction
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PBS:
-
Phosphate buffered saline
References
Allgood AG, Barrick D (2011) J Mol Biol 414(2):243–259
Chung AS, Guan YJ, Yuan ZL, Albina JE, Chin YE (2005) Mol Cell Biol 25(11):4716–4726
De Sepulveda P, Ilangumaran S, Rottapel R (2000) J Biol Chem 275:14005–14008
Debrincat MA, Zhang JG, Willson TA, Silke J, Connolly LM, Simpson RJ, Alexander WS, Nicola NA, Kile BT, Hilton DJ (2007) J Biol Chem 282:4728–4737
Edwards MS, Sternberg JE, Thornton JM (1987) Protein Eng 1:173–181
Ehebauer MT, Chirgadze DY, Hayward P, Martinez Arias A, Blundell TL (2005) Biochem J 392(Pt 1):13–20
Fei X, Zhang Y, Gu X, Qiu R, Mao Y, Ji C (2009) Protein Pept Lett 16:333–335
Gouet P, Courcelle E, Stuart DI, Metoz F (1999) Bioinformatics 15:305–308
Kaddurah-Daouk R, Lillie JW, Daouk GH, Green MR, Kingston R, Schimmel P (1990) Mol Cell Biol 10:1476–1483
Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, Kitamura T, Kato H, Nakayama K, Yoshimura A (2001) J Biol Chem 276:12530–12538
Kile BT, Viney EM, Willson TA, Brodnicki TC, Cancilla MR, Herlihy AS, Croker BA, Baca M, Nicola NA, Hilton DJ, Alexander WS (2000) Gene 258:31–41
Kohroki J, Nishiyama T, Nakamura T, Masuho Y (2005) FEBS Lett 579:6796–6802
Kwon S, Kim D, Rhee J, Park J, Kim D, Kim D, Lee Y, Kwon H (2010) BMC Biol 8:23
Manjasetty BA, Quedenau C, Sievert V, Büssow K, Niesen F, Delbrück H, Heinemann U (2004) Proteins 55(1):214–217
McArthur MW, Laskowski RA, Moss DS, Thornton JM (1993) J Appl Cryst 26:283–291
Michaely P, Tomchick DR, Machius M, Anderson RG (2002) EMBO J 21:6387–6396
Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z (2004) Protein Sci 13:1435–1448
Murshudov GN, Vagin AA, Dodson EJ (1997) Acta Crystallogr D 53:240–255
Notredame C, Higgins DG, Heringa J (2000) J Mol Biol 302:205–217
Sedgwick SG, Smerdon SJ (1999) Trends Biochem Sci 24:311–316
Stebbins CE, Kaelin WG, Pavletich NP (1999) Science 284:455–461
Storoni LC, McCoy AJ, Read RJ (2004) Acta Crystallogr D Biol Crystallogr 60:432–438
Wallimann T, Hemmer W (1994) Mol Cell Biochem 133:193–220
Wallimann T, Wyss M, Brdiczka D, Nicolay K, Eppenberger HM (1992) Biochem J 281:21–40
Wyss M, Kaddurah-Daouk R (2000) Physiol Rev 80:1107–1213
Zhao J, Schmieg FI, Simmons DT, Molloy GR (1994) Mol Cell Biol 14:8483–8492
Zweifel ME, Barrick D (2001) Biochemistry 40(48):14344–14356
Zweifel ME, Barrick D (2001) Biochemistry 40(48):14357–14367
Zweifel ME, Leahy DJ, Hughson FM, Barrick D (2003) Protein Sci 12(11):2622–2632
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program, 2007CB914304 & 2009CB825505), the National Natural Science Foundation of China (30770427), the New Century Excellent Talents in University (NCET-06-0356), the Shanghai Leading Academic Discipline Project (B111) and the National Talent Training Fund in Basic Research of China (No. J0630643).
Author information
Authors and Affiliations
Corresponding author
Additional information
The crystal structure reported in this paper has been submitted to the PDB with ID code 3D9H.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fei, X., Gu, X., Fan, S. et al. Crystal Structure of Human ASB9-2 and Substrate-Recognition of CKB. Protein J 31, 275–284 (2012). https://doi.org/10.1007/s10930-012-9401-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-012-9401-1