Abstract
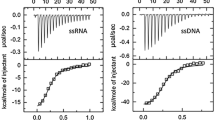
Single-stranded DNA-binding protein (SSB) plays an important role in DNA metabolism, such as DNA replication, repair, and recombination, and is essential for cell survival. We characterized the single-stranded DNA (ssDNA)-binding properties of Salmonella enterica serovar Typhimurium LT2 SSB (StSSB) by using fluorescence quenching measurements and electrophoretic mobility shift analysis (EMSA). Analysis of purified StSSB by gel filtration chromatography showed a stable tetramer in solution. In fluorescence titrations, StSSB bound to 21–38 nucleotides (nt) per tetramer depending on the salt concentration. Using EMSA, we characterized the stoichiometry of StSSB complexed with a series of ssDNA homopolymers, and the size of the binding site was determined to be 22 ± 1 nt. Furthermore, EMSA results indicated that the dissociation constants of StSSB for the first tetramer were less than that for the second tetramer. On the basis of these biophysical analyses, the ssDNA binding-mode of StSSB is expected to be noncooperative.







Similar content being viewed by others
Abbreviations
- Ec :
-
Escherichia coli
- St :
-
Salmonella enterica serovar Typhimurium LT2
- Pa :
-
Pseudomonas aeruginosa PAO1
- Mt :
-
Mycobacterium tuberculosis
- Ms :
-
Mycobacterium smegmatis
- Hp :
-
Helicobacter pylori
- ssDNA:
-
Single-stranded DNA
- SSB:
-
Single-stranded DNA-binding protein
- SDS-PAGE:
-
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis
- EDTA:
-
Ethylenediamine tetraacetic acid
- EMSA:
-
Electrophoretic mobility shift analysis
- nt:
-
Nucleotides
- K d :
-
The apparent dissociation constant
References
Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ (2009) J Mol Biol 388:508–519
Chen CC, Huang CY (2011) Protein J. doi:10.1007/s10930-010-9302-0
Curth U, Genschel J, Urbanke C, Greipel J (1996) Nucleic Acids Res 24:2706–2711
Curth U, Greipel J, Urbanke C, Maass G (1993) Biochemistry 32:2585–2591
Dabrowski S, Olszewski M, Piatek R, Brillowska-Dabrowska A, Konopa G, Kur J (2002) Microbiology 148:3307–3315
Fairall L, Buttinelli M, Panetta G (2000) Bandshift, gel retardation or electrophoretic mobility shift assays. In: Travers A, Buckle M (eds) DNA-protein interactions: a practical approach. Oxford University Press, New York, pp 65–74
Fedorov R, Witte G, Urbanke C, Manstein DJ, Curth U (2006) Nucleic Acids Res 34:6708–6717
Haseltine CA, Kowalczykowski SC (2002) Mol Microbiol 43:1505–1515
Hsieh HC, Huang CY (2011) Biochem Biophys Res Commun 404:546–551
Huang CY, Chang YW, Chen WT (2008) Biochem Biophys Res Commun 375:220–224
Huang CY, Hsu CH, Sun YJ, Wu HN, Hsiao CD (2006) Nucleic Acids Res 34:3878–3886
Jan HC, Lee YL, Huang CY (2011) Protein J doi:10.1007/s10930-010-9297-6
Kerr ID, Wadsworth RI, Cubeddu L, Blankenfeldt W, Naismith JH, White MF (2003) EMBO J 22:2561–2570
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Bioinformatics 23:2947–2948
Liu JH, Chang TW, Huang CY, Chen SU, Wu HN, Chang MC, Hsiao CD (2004) J Biol Chem 279:50465–50471
Lohman TM, Ferrari ME (1994) Annu Rev Biochem 63:527–570
Madden TL, Tatusov RL, Zhang J (1996) Methods Enzymol 266:131–141
Matos RG, Barbas A, Arraiano CM (2010) Protein J 29:394–397
McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK (2001) Nature 413:852–856
Meltzer E, Schwartz E (2010) Curr Opin Infect Dis 23:432–437
Murzin AG (1993) EMBO J 12:861–867
Naue N, Fedorov R, Pich A, Manstein DJ, Curth U (2010) Nucleic Acids Res. doi:10.1093/nar/gkq988
Oakley GG, Patrick SM (2010) Front Biosci 15:883–900
Olszewski M, Grot A, Wojciechowski M, Nowak M, Mickiewicz M, Kur J (2010) BMC Microbiol 10:260
Olszewski M, Mickiewicz M, Kur J (2008) Arch Microbiol 190:79–87
Raghunathan S, Kozlov AG, Lohman TM, Waksman G (2000) Nat Struct Biol 7:648–652
Reyes-Lamothe R, Sherratt DJ, Leake MC (2010) Science 328:498–501
Roy R, Kozlov AG, Lohman TM, Ha T (2007) J Mol Biol 369:1244–1257
Roy R, Kozlov AG, Lohman TM, Ha T (2009) Nature 461:1092–1097
Saikrishnan K, Jeyakanthan J, Venkatesh J, Acharya N, Sekar K, Varshney U, Vijayan M (2003) J Mol Biol 331:385–393
Saikrishnan K, Manjunath GP, Singh P, Jeyakanthan J, Dauter Z, Sekar K, Muniyappa K, Vijayan M (2005) Acta Crystallogr D Biol Crystallogr 61:1140–1148
Savvides SN, Raghunathan S, Futterer K, Kozlov AG, Lohman TM, Waksman G (2004) Protein Sci 13:1942–1947
Schwarz G, Watanabe F (1983) J Mol Biol 163:467–484
Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL (2008) Crit Rev Biochem Mol Biol 43:289–318
Tischler AD, McKinney JD (2010) Curr Opin Microbiol 13:93–99
Tsolis RM, Young GM, Solnick JV, Baumler AJ (2008) Nat Rev Microbiol 6:883–892
Wadsworth RI, White MF (2001) Nucleic Acids Res 29:914–920
Wang CC, Tsau HW, Chen WT, Huang CY (2010) Protein J 29:445–452
Witte G, Fedorov R, Curth U (2008) Biophys J 94:2269–2279
Witte G, Urbanke C, Curth U (2003) Nucleic Acids Res 31:4434–4440
Witte G, Urbanke C, Curth U (2005) Nucleic Acids Res 33:1662–1670
Wold MS (1997) Annu Rev Biochem 66:61–92
Acknowledgments
We thank Ms. Hui-Chuan Hsieh for constructing the pET21b-StSSB. This research was supported a grant from the National Research Program for Genome Medicine (NSC 99-3112-B-040-001 to C.Y. Huang).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, YH., Lee, YL. & Huang, CY. Characterization of a Single-Stranded DNA Binding Protein from Salmonella enterica Serovar Typhimurium LT2. Protein J 30, 102–108 (2011). https://doi.org/10.1007/s10930-011-9309-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-011-9309-1